This package is a library of UI components:
- to display Feature Table objects in appropriate table
- to display Sequence Information objects in appropriate viewer
- to display DNA and Protein Sequence objects in various viewers: text, graphic, HCA (protein only), etc.
- to display BLAST result objects in an appropriate viewer
In other words, this library is the appropriate UI friend of the Bioinformatics Core library.
Here is an example of use (more, below):
Use a Java Virtual Machine 1.8 (or above) from Oracle.
Not tested with any other JVM providers but Oracle... so there is no guarantee that the software will work as expected if not using Oracle's JVM.
Since this package is a library, its primary purpose targets a use within other softwares. The package contains a quite large number of classes, most of them being actually supporting code to these viewers:
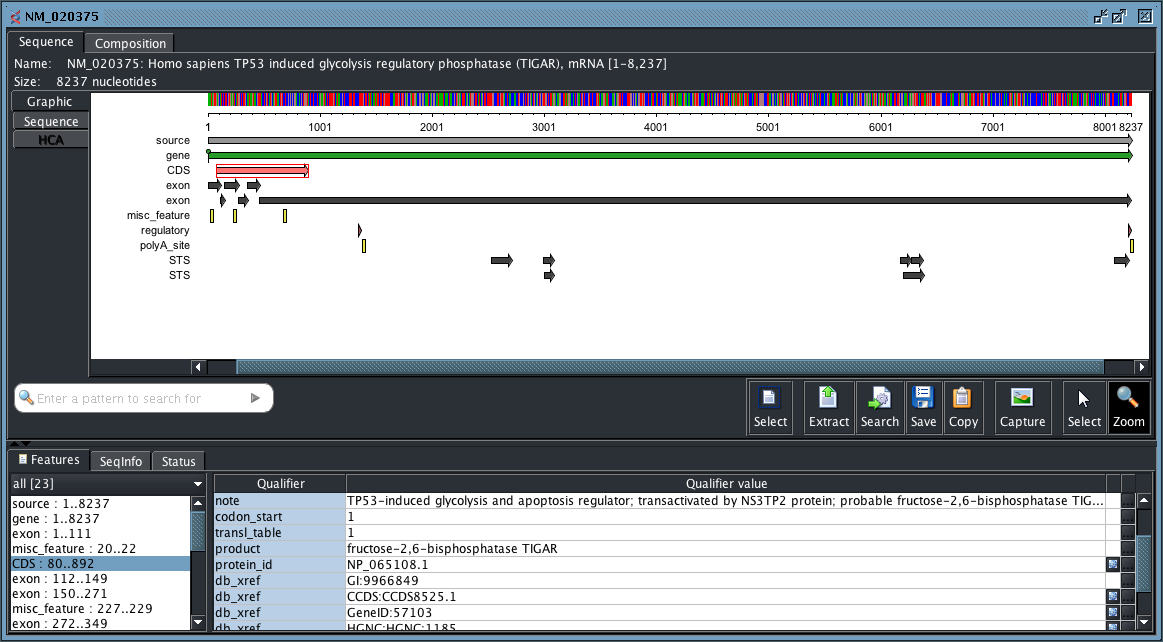
- FeatureViewer: a viewer for FeatureTable objects
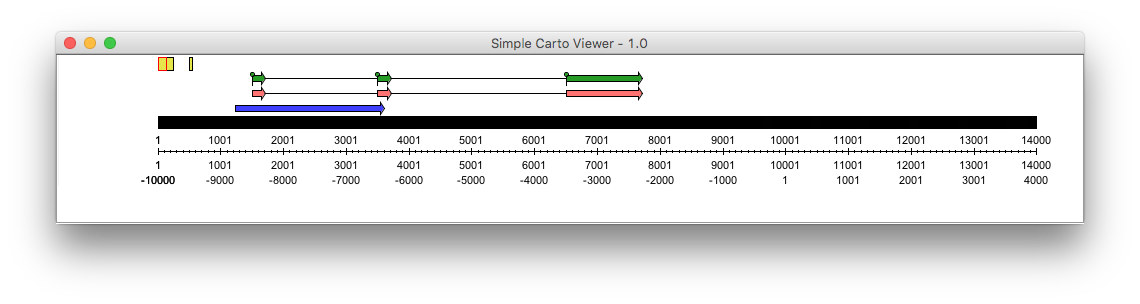
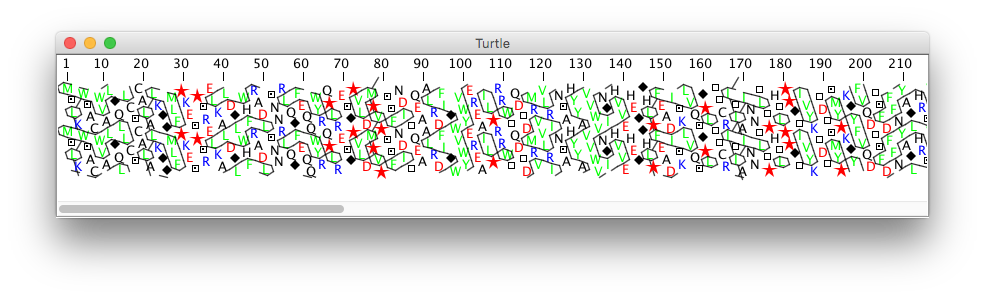
- CartoViewerPanel: a graphical sequence viewer
- PanelHCA: a protein sequence viewer relying on Hydrophobic Cluster Analysis (HCA)
- DSequenceTableViewer: a sequence viewer relying on a table
- AlphabetCounterXXXViewer: display letter content of a sequence (i.e. its composition)
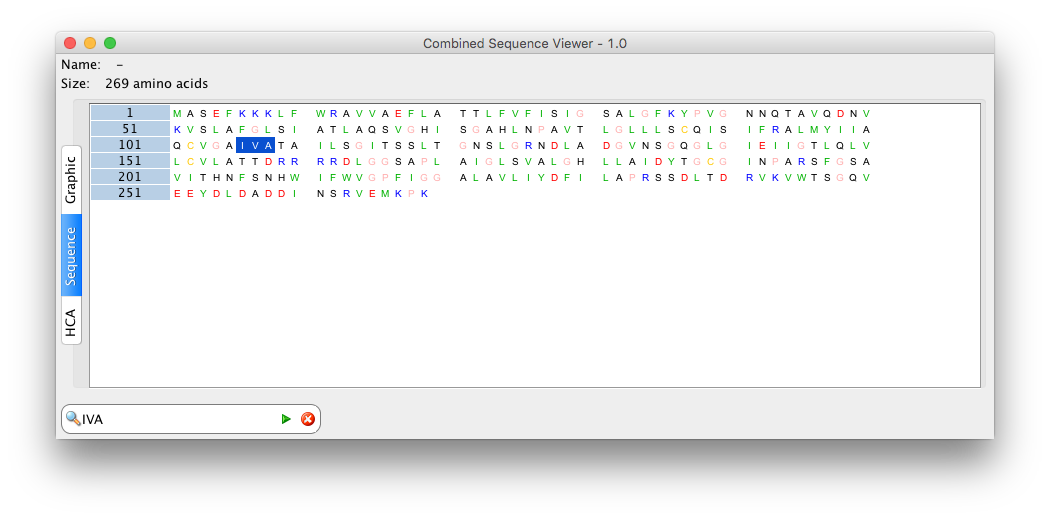
- CombinedSequenceViewer: combine viewers 2, 3 and 4 into a single one
- CombinedSequenceViewerExt: combine viewers 2, 3, 4 and 5 into a single one
- CombinedAnnotatedSequenceViewer: everything in a single full-featured viewer (above picture)
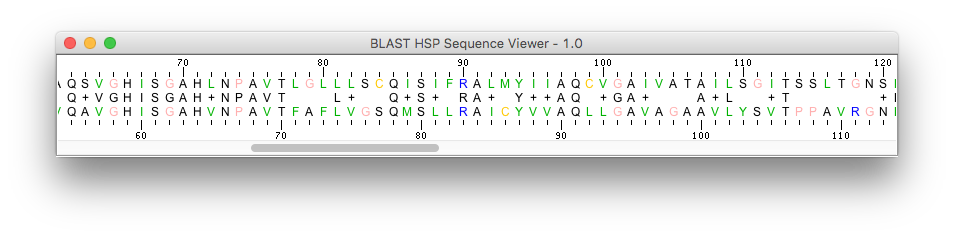
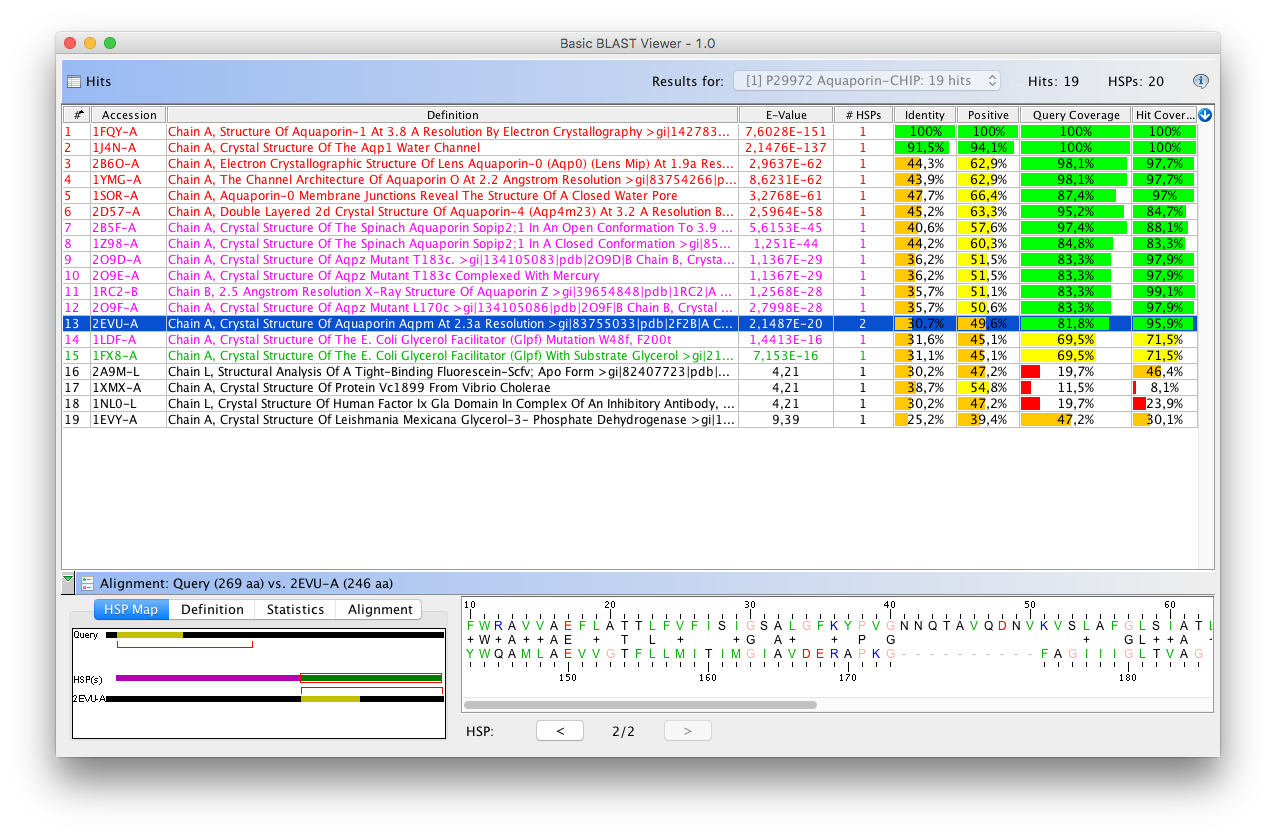
- BlastViewerPanelBase: a graphical BLAST results viewer component
You can see how to use the library by having a look at the "example" source package. It contains sample source codes to illustrate the use of these components:
: see how to create a simple sequence viewer
: see how to load and display BLAST pairwise sequence alignments
: see how to create a graphic representation of a sequence
: see to create a combined sequence viewer
: see how to create a HCA view of a protein sequence
: see how to create a BLAST Viewer tool
See also Wiki.
Have in mind that this library can be a nice friend to Biojava: Sequence objects from Biojava can be converted quite easily to Sequence objects from Bioinformatics Core library for the purpose of using this Bioinformatics UI library. See Biojava Convertor Project to review how to do that.
Bioinformatics UI API itself is released under the GNU Affero General Public License, Version 3.0. AGPL
It depends on several thrid-party libraries as stated in the NOTICE.txt file provided with this project.
During Summer 2004 I was asking myself how easy it could be to rewrite in Java the original VisualBlast software written in C for the MS-Windows platform.
Actually, the answer was quite immediate: it took me a few days to fully rewrite Visual Blast, thanks to the many libraries available, mostly from Sun MicroSystems and the Apache Software Foundation.
So, during my holidays on Summer 2004, I started a wonderful story: the creation of what would eventually become the KoriBlast/ngKLAST suite of softwares, the founding software of my company, Korilog.
Korilog ceased its activities on June 2015, therefore I decided to release these UI components to the open source community.
-- (c) 2004-2023 - Patrick G. Durand