Python tools for TCR:peptide-MHC modeling and analysis
Described in the manuscript: "Structure-based prediction of T cell receptor:peptide-MHC interactions" by Philip Bradley. bioRXiv link:
[NEW 2024-03-06] The AlphaFold code was updated to alphafold v2.3.2 from alphafold repo commit f715f016d875e2fd8515457af2b12277a716b9b0 The old code can be found in the releases section under v1.0.0
[NEW 2024-01-17] The Google colab notebook was temporarily broken due to an update of the colab GPU libraries (CUDA/CUDNN). I just checked in an updated version that should be working again. This is a short-term patch until I update the main branch to use the latest AlphaFold code.
[NEW 2023-10-01] Google colab notebook for
running TCR:pMHC structure predictions
with an updated, fine-tuned model and streamlined workflow (3x faster simulations
than in the paper), which you can
- Set up and run TCR-specialized AlphaFold simulations starting from a TSV file with TCR, peptide, and MHC information.
- Parse a TCR:peptide-MHC ternary PDB structure and define V/J/CDR3, MHC allele, TCR and MHC coordinate frames, and TCR:pMHC docking geometry
- Calculate distances between docking geometries ('docking RMSDs') for use in clustering/docking analysis and model evaluation.
This functionality is made available in a Python package (tcrdock) and through
wrapper scripts as illustrated in the examples below.
The tcrdock package includes a subset of the
TCRdist library as a submodule (tcrdock/tcrdist) for use during the AlphaFold
setup process. Refer to the source of the compute_tcrdists.py script for a
simple use case.
NOTE: Input and output sequence/residue numbers are 0-indexed:
they start at 0 and run
through N-1, where N is the length of the sequence. For example, the MHC and TCR
core positions written out by parse_tcr_pmhc_pdbfile.py are 0-indexed with
respect to the full ternary sequence.
Running any of these Python scripts with the -h flag should print a help mesage.
The PDB file should just contain a single copy of the ternary structure, with the following chains (in order)
MHC-I: (1) MHC, (2) beta-2 microglobulin, (3) peptide, (4) TCRA, (5) TCRB
or MHC-I: (1) MHC, (2) peptide, (3) TCRA, (4) TCRB
MHC-II: (1) MHCA, (2) MHCB, (3) peptide, (4) TCRA, (5) TCRB
If you run into trouble, it's not a bad idea to "clean" the pdb file by removing extraneous ligands/waters/etc.
python parse_tcr_pmhc_pdbfile.py --pdbfiles examples/parsing/1qsf.pdb \
--organism human --mhc_class 1 --out_tsvfile parse_output.tsv
python setup_for_alphafold.py --targets_tsvfile examples/benchmark/single_target.tsv \
--output_dir test_setup_single
or
python setup_for_alphafold.py --targets_tsvfile examples/benchmark/full_benchmark.tsv \
--output_dir test_setup_full_benchmark --benchmark
Here the --benchmark flag tells the script to exclude nearby templates.
Here $ALPHAFOLD_DATA_DIR should point to a folder that contains the AlphaFold
params/ folder.
This will use the very-slightly-modified version of the alphafold library included
with this repository (see changes_to_alphafold.txt). It should also run OK
with any post-Nov 2021 version of
alphafold, but it may not be as efficient (length changes in the targets list
will trigger recompilation of the model). You will need to run in a Python environment
that has the packages required by alphafold (or in the alphafold docker instance).
If you have an older (pre-multimer) version of alphafold, try changing the variable
NEW_ALPHAFOLD to False at the top of predict_utils.py.
python run_prediction.py --targets test_setup_single/targets.tsv \
--outfile_prefix test_run_single --model_names model_2_ptm \
--data_dir $ALPHAFOLD_DATA_DIR
or
python run_prediction.py --targets test_setup_full_benchmark/targets.tsv \
--outfile_prefix test_run_full --model_names model_2_ptm \
--data_dir $ALPHAFOLD_DATA_DIR
This will compute the matrix of docking RMSDs among the 220 ternary TCR:pMHC complex
structures in the current template database using the info stored in the file
tcrdock/db/ternary_templates_v2.tsv.
python compute_docking_rmsds.py --docking_geometries_tsvfile tcrdock/db/ternary_templates_v2.tsv \
--outfile rmsds.txt
This will compute the matrix of paired TCRdist values and save it to a text file.
It will also print out the TCRdiv repertoire diversity measure calculated
with the sigma from the command line flag --tcrdiv_sigma (the default is 120)
python compute_tcrdists.py --tcrs_tsvfile examples/tcrdist/human_tcrs.tsv \
--organism human --outfile tcrdists.txt
The non-AlphaFold Python package requirements are listed in requirements.txt.
Those specific
package versions should work, but there should also be plenty of flexibility on the
versions. The TCR and MHC parsing code also requires the NCBI BLAST+ software
to be installed, which can be done by running the script
download_blast.py. A potential installation route would be:
conda create --name tcrdock_test python=3.8
source activate tcrdock_test # or: conda activate tcrdock_test
pip3 install -r requirements.txt
python download_blast.py
To run the AlphaFold simulations, you will need a Python environment that satisfies additional requirements as explained further in the AlphaFold README. One option would be to use the AlphaFold Docker container.
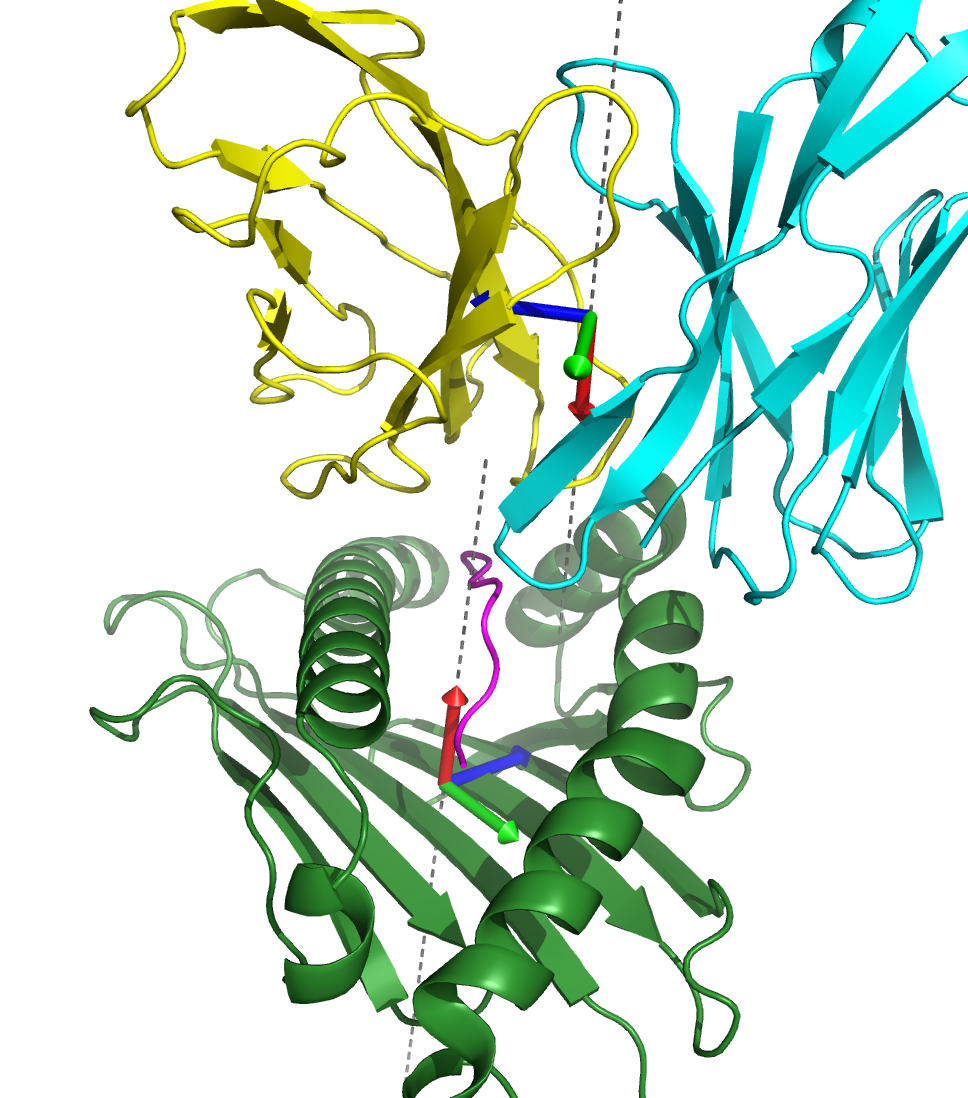
TCR:pMHC docking geometries represent the coordinate transformation between
the MHC and TCR reference frames, which are shown as red (X-axis), green (Y-axis), and
blue (Z-axis) arrow triplets in the above TCR:peptide-MHC structure. The
tcrdock.docking_geometry.DockingGeometry class defined in
tcrdock/docking_geometry.py stores docking geometry information.
Docking geometries are represented (internally and in parsing output)
by 6 numbers and two booleans. The exact choice of which 6 numbers to use to
represent this rigid body transformation is guided by the fact that
the MHC X-axis points up toward the peptide and toward the "canonical"
TCR location, and the TCR X-axis points from near the TCR center of mass
along the TCRA/TCRB symmetry axis toward the CDR3 loops and toward the
"canonical" MHC location (see red arrows in image above).
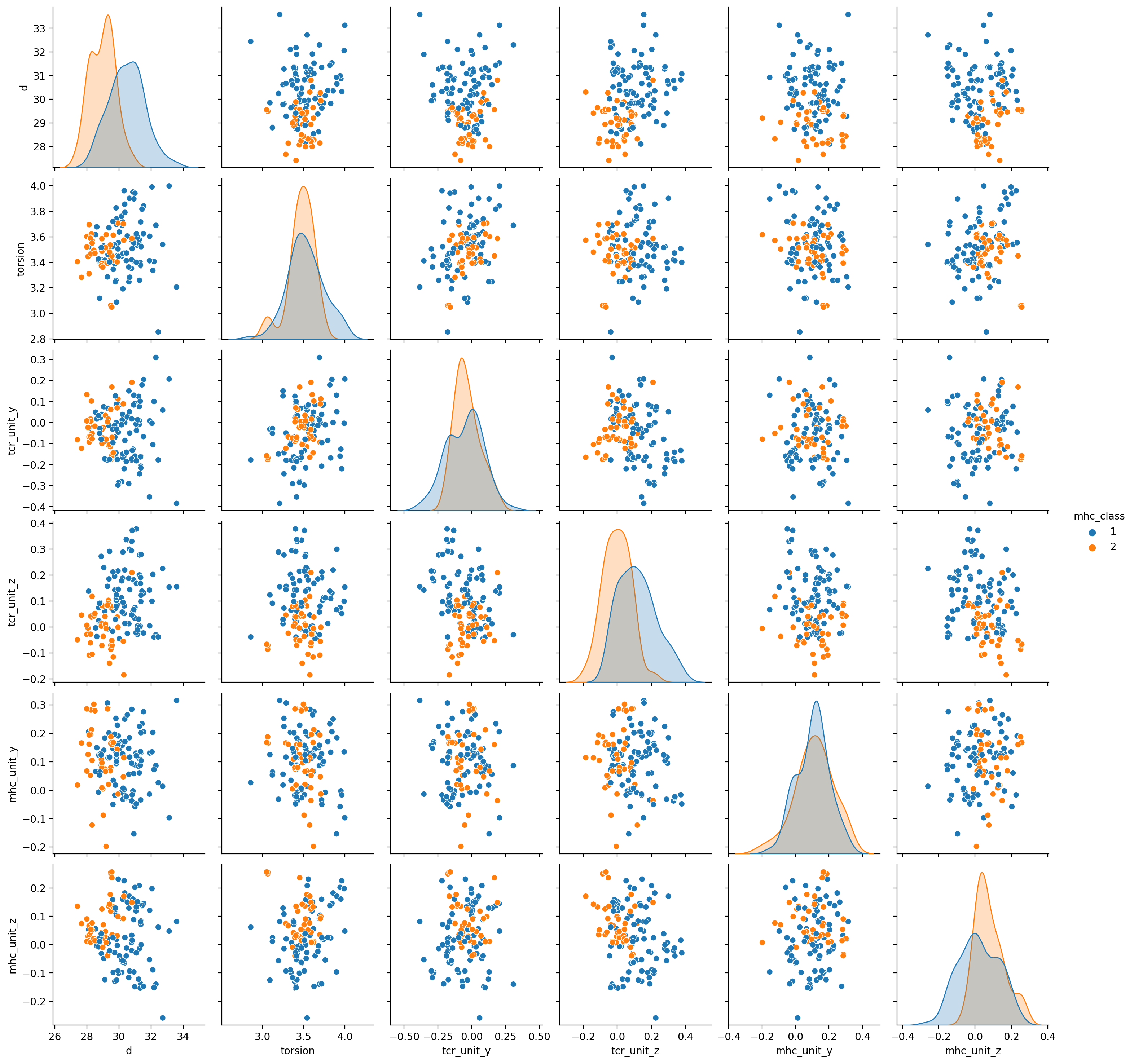
The docking geometry parameters are as follows (see below for scatterplots):
-
d: the distance in Angstroms between the MHC and TCR coordinate frame origins.
-
torsion: the dihedral angle in radians between the MHC Y-axis and the TCR Z-axis, (typically around pi). Takes values in the interval [0,2*pi)
-
tcr_unit_y: The Y-component of the unit vector that points from MHC to TCR, in the MHC frame. In other words, this is the location of the TCR, written in the MHC coordinate frame, normalized to have length 1. This vector is usually pretty close to the X-axis vector, since the X-axis of the MHC frame points up from the beta sheet center toward the peptide, and is perpendicular to the beta sheet.
-
tcr_unit_z: The Z-component of the unit vector that points from MHC to TCR, in the MHC frame.
-
mhc_unit_y: The Y-component of the unit vector that points from TCR to MHC, in the TCR frame. In other words, this is the location of the MHC, written in the TCR coordinate frame, normalized to have length 1. This vector is usually pretty close to the X-axis vector, since the X-axis of the TCR frame points along the TCRA/TCRB symmetry axis toward the CDR3 loops.
-
mhc_unit_z: The Z-component of the unit vector that points from TCR to MHC, in the TCR frame.
-
tcr_unit_x_is_negative: boolean that tells whether the MHC--->TCR unit vector has a negative x-component. This is always
Falsefor canonical docking geometries. It would beTrueif the TCR is docking to the underside of the MHC beta sheet, or if the peptide is underneath the beta sheet (ie, whenever TCR and peptide are on opposite sides of the MHC beta sheet). Assumed to beFalseif absent from the inputs. -
mhc_unit_x_is_negative: boolean that tells whether the TCR--->MHC unit vector has a negative x-component. This is always
Falsefor canonical docking geometries. It would beTrueif the TCR CDR3 loops are pointing away from the MHC, for example if the TCR is interacting with the MHC via the constant domain. Assumed to beFalseif absent from the inputs.
To see the details of the calculation, refer to the DockingGeometry.from_stubs
function. Currently it's here:
https://github.com/phbradley/TCRdock/blob/main/tcrdock/docking_geometry.py#L47
Here are some scatter plots of docking geometry parameters for the 130 ternary structures in the TCR structure prediction benchmark.
The input TSV file for the setup_for_alphafold.py script should have the 10 columns
organism, mhc_class, mhc, peptide, va, ja, cdr3a, vb, jb, cdr3b
More explanation can be found by running python setup_for_alphafold.py -h
The format of the targets and alignments files created by the setup_for_alphafold.py
script and read by the run_prediction.py script are explained in the
README for
the alphafold_finetune repository (from which that script is borrowed).