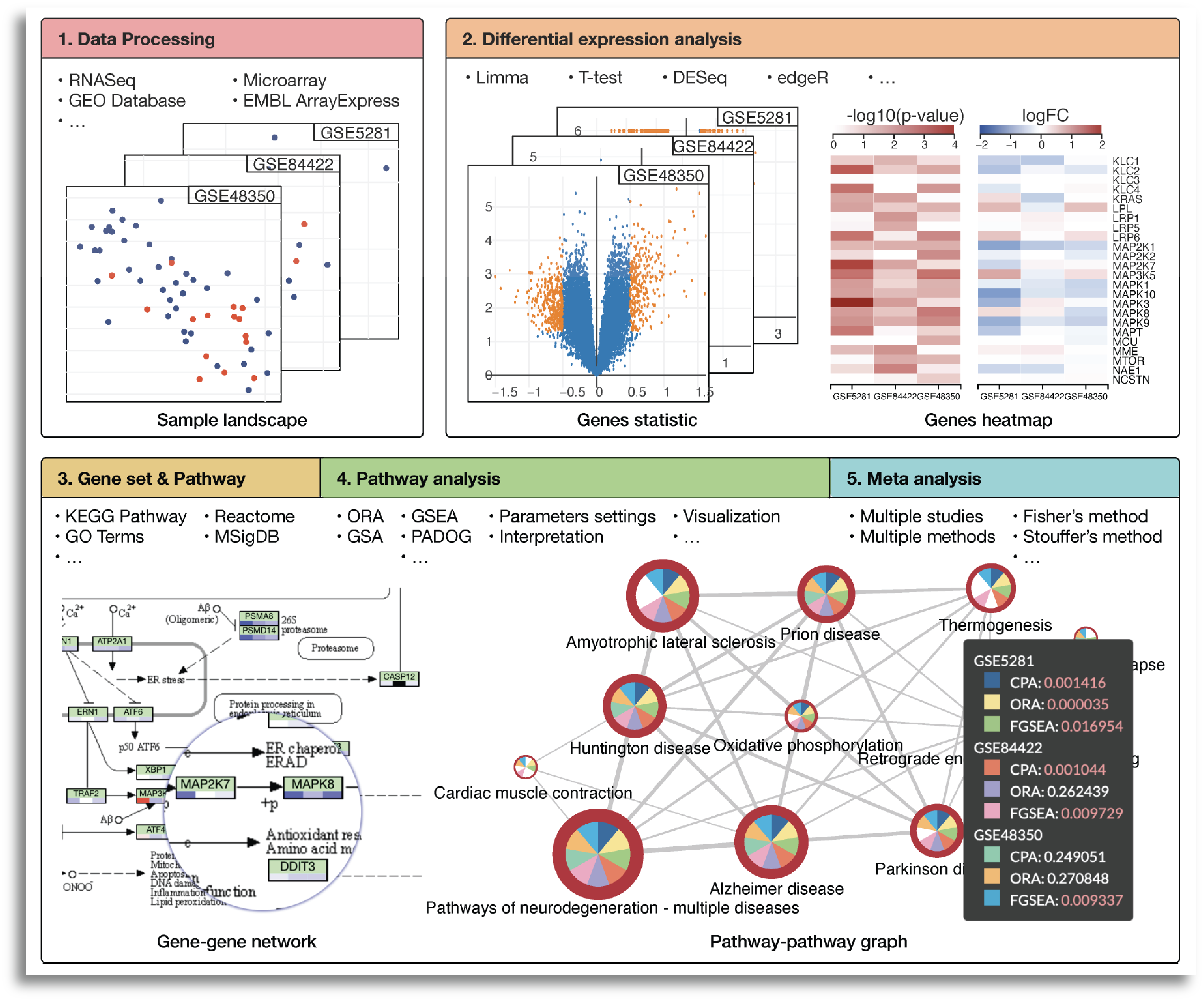
This cloud-based learning module is specialized for pathway analyses using omics datasets to discover consistent biological mechanism behind a condition. The content will be arranged in five sub-modules which allows us to:
- Download and process data from public repositories
- Perform differential analysis
- Perform pathway analysis using different methods that seek to answer different research hypotheses
- Perform meta-analysis and combine methods and datasets to find consensus results
- Interactively explore significantly impacted pathways across multiple analyses, and browsing relationships between pathways and genes.
Each learning sub-modules will be organized in a R Jupyter notebook to help the participants familiarize themselves with the cloud computing in the specific context of running bioinformatics workflows. Each notebook will include step-by-step hand-one practice with R command line to install necessary tools, obtain data, perform analysis, visualize and interpret the results.
This learning module does require any computational hardware and local environment setting from users. However, users need to have Google email account, sufficient internet access, and a stardard web-browser (e.g. Chrome, Edge, Firefox etc., Chrome browser is recommended) to create a cloud virtual machine for analysis.
The content of the course is organized in R Jupyter Notebook. Then we use Jupyter Book which is a package to combine individuals Jupyter Notebooks into a web-interface for a better navigation. Details of installing the tools and formatting the content can be found at: https://jupyterbook.org/en/stable/intro.html. The content of the course is deposited to Dr. Tin Nguyen Github repository at https://github.com/tinnlab/NOSI-Google-Cloud-Training.
The Jupyter Book will run and test all codes in our notebooks and then build them into a website format.
The website output is the _build/html folder in the repository. After the _build/html is completely built, the web version
of the book will be uploaded the _build/html folder to a Google bucket.
The address of the cloud bucket can be found at: https://storage.googleapis.com/nosi-gcloud-course/html/intro.html.
The link http://tinnguyen-lab.com/course/cpa-gcloud/intro.html is just a proxy to that link using Dr. Tin Nguyen domain
name.
A completed workflow from setting up the environment, building the book, to uploading the website to Google cloud bucket here: https://github.com/tinnlab/NOSI-Google-Cloud-Training/blob/main/.github/workflows/main.yml. The logs for each time it builds the book are here: https://github.com/tinnlab/NOSI-Google-Cloud-Training/actions