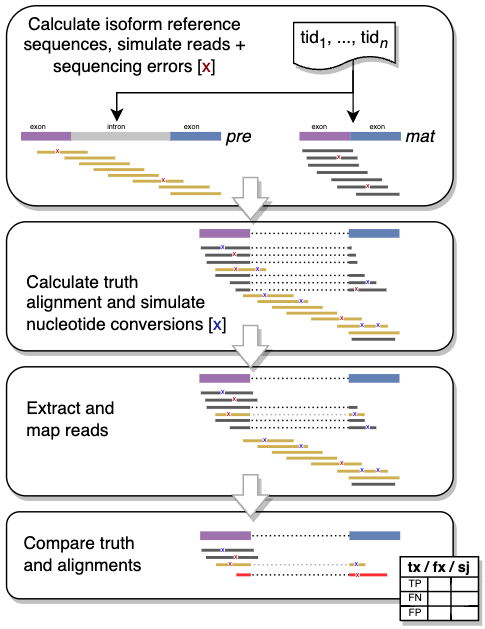
splice_sim is a python based RNA-seq simulation and evaluation package specialized on the simulation nucleotide-conversions and splice isoforms. We use Nextflow and containerization via Docker to wrap splice_sim into a readily executable start-to-end framework for simulating and evaluating complex experimental scenarios with mapping based processing pipelines.
- Realistic Illumina short-read simulation using ART
- Simulation of customizable nucleotide-conversions at configurable rates
- Simulation of isoforms at configurable splicing states per transcript
- Mapping accuracies and performance metrics at different scopes of genomic annotation
- Elaborate output tracks for visual inspection stratified by performance metric
splice_sim itself is a python package with several dependencies that are best simply installed in a conda environment:
conda env create -f environment.ymlThen clone the splice_sim repository and call its main method:
git clone https://github.com/popitsch/splice_sim.git
cd splice_sim
python main.pyTo run our full-blown all-on-one Nextflow based workflow, you simply need to install Nextflow and Docker to have all dependencies available and be ready to go. To run splice_sim on HPC environments, most administrators prefer Singularity (apptainer) containers which can be seamlessly created from our Docker container.
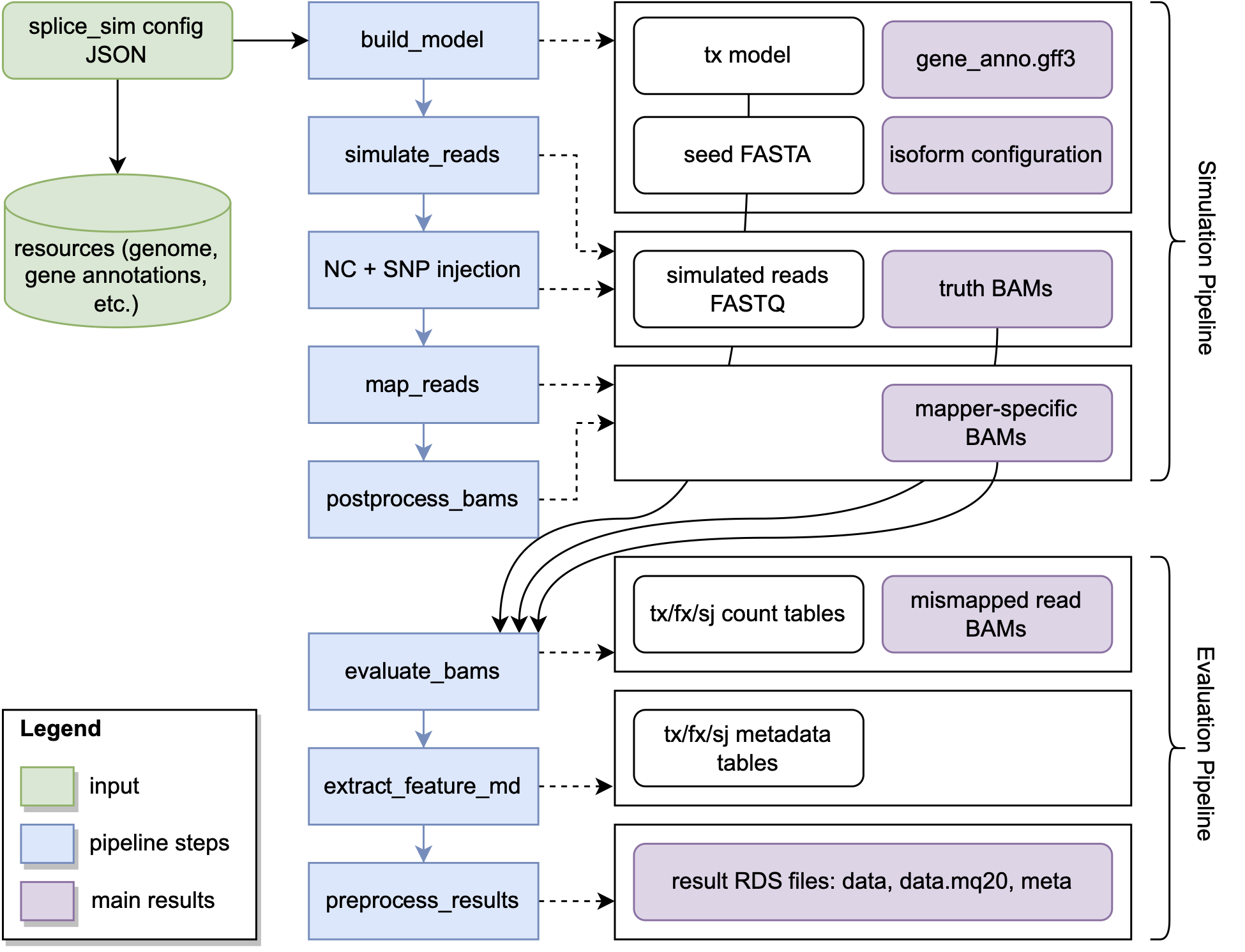
splice_sim itself is the Python package and includes the simulation engine to both simulate and evaluate datasets. splice_sim provides dedicate commands for the individual logical steps outlined below, starting from creating transcript models, simulating reads from those transcripts and finally evaluating the performance of a given mapper via the produced bam file. The parameters for the highlighted relevant steps for such a process are described in the splice_sim engine.
We have also wrapped a ready-to-use out of the box workflow that executes all step from start to end into our Nextflow workflow with a description provided in the adjacent section. We provide two separate workflows: (i) splice_sim.nf that handles the entire logic contained in the Simulation Pipeline block and (ii) splice_sim_eva.nf that handles everything related to the Evaluation Pipeline block in the diagram below. To avoid supplying an extensive set of parameters to theindividual steps via the command line, we designed splice_sim to be driven by a configuration file in json format that contains all necessary inputs and parameters needed for any of splice_sim steps.
All you need to get started with our workflows is Nextflow and Docker. Typically one would also need to tweak the resource requirements for each of the processes that are executed. We did that already for you for the small (typically expressed genes size) and large (full transcriptome) datasets simulated in our study in the provided nextflow.config.example ready to use. Both workflows need as only input the configuration file file provided by the user.
This workflow executes the following steps to obtain the simulated truth alignments and mapped read alignments of the mappers under investigation in BAM format as final output:
- Build a transcript model with
splice_sim build_model - Simulate reads with ART for all conditions
- Calculate and encode truth alignments for all conditions
- Map simulated reads with mappers under investigation for all conditions
- Postprocess BAM files with
splice_sim postfilter_bam
This workflow executes the following steps to obtain the evaluation metrics and output files from the simulated truth alignments and mapped read alignments of the mappers under investigation as final output:
- Evaluate a given read alignment of a mapper with
splice_sim evaluate - Extract associated metadata of the annotated features under investigation with
splice_sim extract_feature_metadata - Bundle, package and compress the produced output tables into RDS files to futher process in R with
preprocess_results.R
The core splice_sim python engine covers isolated steps of the simulation and evaluation process process as highlighted in the previous block diagram. The most important commands along the way a documented in this section.
The build_model command takes the reference and configuration provided by the user and creates the transcript model and sequence files needed that contains the composition of the transcriptome and serves and input to the read simulation step of splice_sim.
python splice_sim/main.py build_model --help
usage: main.py [-h] -c config_file [-o outdir]
Copyright (C) 2021 XXX. All rights reserved.
Distributed on an "AS IS" basis without warranties
or conditions of any kind, either express or implied.
USAGE
optional arguments:
-h, --help show this help message and exit
-c config_file, --config config_file
JSON config file
-o outdir, --outdir outdir
output directory (default is current dir)The create_genome_bam command takes the simulated read set with your short-read simulator of choice (we use ART) and calculates the truth alignments that serve as the reference benchmark in splice_sim.
python splice_sim/main.py create_genome_bam --help
usage: main.py [-h] -m model_file -a config_file [-t threads] [-o outdir]
Copyright (C) 2021 XXX. All rights reserved.
Distributed on an "AS IS" basis without warranties
or conditions of any kind, either express or implied.
USAGE
optional arguments:
-h, --help show this help message and exit
-m model_file, --model model_file
model file
-a config_file, --art_sam config_file
ART sam file
-t threads, --threads threads
threads
-o outdir, --outdir outdir
output directory (default is current dir)The postfilter_bam command filters secondary and supplementary reads and highlights isoforms.
python splice_sim/main.py postfilter_bam --help
usage: main.py [-h] -c config_file -b bam_file -o outdir
Copyright (C) 2021 XXX. All rights reserved.
Distributed on an "AS IS" basis without warranties
or conditions of any kind, either express or implied.
USAGE
optional arguments:
-h, --help show this help message and exit
-c config_file, --config config_file
JSON config file
-b bam_file, --bam bam_file
input bam file
-o outdir, --outdir outdir
output dirThe evaluate command runs the splice_sim evaluation routine on a given mapper bam file produced for a condition of the splice_sim simulation run.
python splice_sim/main.py evaluate --help
usage: main.py [-h] -b bam_file -c config_file -m model_file [-f filter_bed] [-t THREADS] -o outdir
Copyright (C) 2021 XXX. All rights reserved.
Distributed on an "AS IS" basis without warranties
or conditions of any kind, either express or implied.
USAGE
optional arguments:
-h, --help show this help message and exit
-b bam_file, --bam_file bam_file
input bam file
-c config_file, --config config_file
JSON config file
-m model_file, --model model_file
model file
-f filter_bed, --filter_bed filter_bed
Filter regions BED file
-t THREADS, --threads THREADS
used threads
-o outdir, --outdir outdir
output dirThe extract_feature_metadata extracts comprehensive metadata that lists various characteristics of the genomic features under evaluation.
python splice_sim/main.py extract_feature_metadata --help
usage: main.py [-h] -c config_file -m model_file -o outdir
Copyright (C) 2021 XXX. All rights reserved.
Distributed on an "AS IS" basis without warranties
or conditions of any kind, either express or implied.
USAGE
optional arguments:
-h, --help show this help message and exit
-c config_file, --config config_file
JSON config file
-m model_file, --model model_file
model file
-o outdir, --outdir outdir
output dirAll the above commands of the splice_sim engine can be run without installing any dependencies out of the box of our Docker container. Simply precede the individual splice_sim calls like illustrated below:
docker run --cpus <number of CPUS> -m <GB memory>g -v $(pwd):$(pwd) -w $(pwd) tobneu/splice_sim:release python <path to splice_sim repository>/main.py
We provide a Rscript that takes all splice_sim evaluation outputs and processes them into readily usable RDS objects to be imported in R. The script is located in splice_sim/src/main/R/splice_sim/preprocess_results.R and all dependencies are again wrapped into our splice_sim R Docker container tobneu/splice_sim_r:latest.
Rscript --vanilla splice_sim/src/main/R/splice_sim/preprocess_results.R
Error: usage: preprocess_results.R <splice_sim_config> [<outdir>]
Execution halted
We made the deliberate decision to have a central configuration file in json format that encapsulate and document all input files and parameters that are driving all splice_sim related processes. Here we document all required and optional parameters:
| Parameter | Description | Required |
|---|---|---|
dataset_name |
Name of the simulation run | ✅ |
splice_sim_cmd |
Path to main.py in the clone splice_sim repository |
✅ |
splice_eva_preprocess_cmd |
Path to main.py in the cloned splice_sim repository |
✅ |
gene_gff |
Path to the transcriptome annotation in gff3-format (obtained e.g. from Gencode | ✅ |
intron_gff |
Annotated introns from the transcriptome annotation in gff3-format | ✅ |
genome_fa |
Reference genome sequence in fasta format | ✅ |
genome_chromosome_sizes |
Chromosome lengths file with chromosome name and chromosome lengths separated by a tab as produced by samtools faidx |
✅ |
genome_conservation |
Genome conservation score in bigwig format (obtained e.g. from UCSC) | ❌ |
genome_mappability |
Genome mappability score in bigwig format (obtained e.g. from the Hoffman lab) | ❌ |
transcript_data |
Transcript state configuration file | ❌ |
transcript_ids |
Subset of transcript IDs from the reference annotation used in this simulation | ✅ |
isoform_mode |
Fraction of unspliced to mature spliced transcripts | ✅ |
frac_old_mature |
Fraction of pre-existing fully-spliced transcripts before labelling onset | ✅ |
condition |
Dictionary of the conditions in this simulation defining the base conversion type (ref and alt base), conversion_rates (list of doubles between 0 and 1) and the base_coverage (integer) |
✅ |
mappers |
Dictionary of dictionary of the mappers employed and evaluated in this simulation defining the name of the mapper, the command how to call it, the mapping index and known splice sites file | ✅ |
create_tdf |
Boolean whether TDF files for track visualization in IGV should be produced | ✅ |
max_ilen |
Integer defining the maximum considered intron length | ✅ |
min_abundance |
Integer defining the minimum abundance value fo a transcript to be considered | ✅ |
random_seed |
Integer seed to keep simulation deterministic | ✅ |
readlen |
Integer defining the short read length | ✅ |
write_reads |
Boolean value defining whether reads should be output to disk or not | ❌ |
write_intron_bam |
Boolean value defining whether intronic reads should be written to BAM file not | ❌ |
pre_exist |
Directory of pre-existing runs to restart the simulation in case a run crashes | ❌ |
snp_file |
Optional VCF file containing SNVs to be injected into the simulated data. The VCF file may contain the following (optional) properties in the INFO section: prob: conversion probability [0; 1] (optional, default:1), strand: strand specificity [NA, +, -] (optional, default:NA), enable_nc: [yes, no] (optional, default: yes) if yes, configured NC are possible at the same position if no SNV was injected | ❌ |
Find below an example config json:
{
"dataset_name": "test_simulation",
"splice_sim_cmd": "python /software/splice_sim/main.py", # should point to your local splice_sim clone
"splice_eva_preprocess_cmd": "Rscript --vanilla /software/splice_sim/src/main/R/splice_sim/preprocess_results.R", # should point to your local splice_sim clone
"gene_gff": "/references/gencode.vM21.gff3.gz", # gene annotation GFF3 file
"genome_fa": "/references/Mus_musculus.GRCm38.dna.primary_assembly.fa",
"genome_chromosome_sizes": "/references/Mus_musculus.GRCm38.dna.primary_assembly.fa.chrom.sizes",
"genome_conservation": "/references/mm10.60way.phastCons60wayEuarchontoGlire.bw", # optional
"genome_mappability": "/references/mm10.k24.umap.bedgraph.gz", # optional
"transcript_data": "data.config.json", # if this file exists,
"transcript_ids": "tids.tsv", # optional (TSV file with one column 'transcript_id' containing all considered transcript ids)
"isoform_mode": "1:1", # mode for creating isoform data; '1:1': mat and pre form as in paper (default), 'from_file': data will be loaded from 'transcript_id','abundance','frac_mature','frac_old_mature' columns in the configured transcript_ids file
"frac_old_mature": 0, # fraction of OLD RNA that was not exposed to a nucleotide analog
"condition": {
"ref": "T", # reference base
"alt": "C", # alternat base
"conversion_rates": [ 0.02, 0.04 ], # list of conversion rates
"base_coverage": 10 # used to calculate simulated coverage per tx;
},
"mappers": {
"STAR": {
"star_cmd": "STAR-2.7.1a",
"star_genome_idx": "star_2.7.1",
"star_splice_gtf": "/indices/gencode.vM21.gtf" # GTF file with known splice sites
},
"HISAT3N": {
"hisat3n_cmd": "hisat-3n",
"hisat3n_idx": "/indices/Mus_musculus.GRCm38.dna.primary_assembly",
"hisat3n_kss": "/indices/gencode.vM21.gtf.hisat2_splice_sites.txt" # TXT file with known splice sites
},
"MERANGS": {
"merangs_cmd": "meRanGs",
"star_cmd": "STAR",
"merangs_genome_idx": "/indices/meRanTK-1.2.1b/BSgenomeIDX",
"merangs_splice_gtf": "/indices/gencode.vM21.gtf" # GTF file with known splice sites
},
"SEGEMEHL": {
"segemehl_cmd": "segemehl.x",
"segemehl_ctidx": "/indices/segemehl/Mus_musculus.GRCm38.dna.primary_assembly.ctidx",
"segemehl_gaidx": "/indices/segemehl/Mus_musculus.GRCm38.dna.primary_assembly.gaidx"
}
},
"create_tdf": true,
"max_ilen": 100000,
"min_abundance": 1,
"random_seed": 1234,
"readlen": 100,
"write_reads": false,
"write_intron_bam": false
}These tables contain the performance metrics for a given mapper in the count/*.counts.tsv.gz files.
| Column | Description | Notes |
|---|---|---|
mapper |
Name of the respective mapper; Supported mappers are STAR, HISAT3N, MERANGS and SEGEMEHL | |
conversion_rate |
Conversion rate between 0 and 1.0 | |
fid |
feature/transcript id | |
true_isoform |
name of the isoform (as configured) the read originates from | |
cv1 |
1 if read contains at least one NC or 0 otherwise | |
cv2 |
1 if read contains at least two NC or 0 otherwise | |
se1 |
1 if read contains at least one simulated sequencing error or 0 otherwise | |
se2 |
1 if read contains at least two simulated sequencing errors or 0 otherwise | |
classification |
Read classification: TP: true positive, FN: false negative: FP_raw: false-positive/not normalised, FP: false-positive/normalised |
|
count |
read count. FP classified rows may include fractions | |
class_type |
read type: acc: acceptor spanning, don: donor spanning, spl: spliced read | SJ only |
These tables contain various metadata for the genomic intervals under investigation stratified at transcript level (tx), exon / intron feature level (fx) or splice-junction level (sj) in the meta/*.metadata.tsv.gz files.
| Column | Description | Notes |
|---|---|---|
tid |
Transcript ID | |
fid |
Feature ID (intron or exon ID) | FX+SJ only |
ftype |
Feature type: tx, fx, don, acc or spl |
|
rnk |
Rank. For transcripts this is the number exons, for introns/exons it is the rank from the transcript 5'-end | |
chromosome |
Chromosome of the annotate feature | |
start / end |
Genomic start/end position of the annotated feature | |
strand |
Strand of the annotation | |
A/C/T/G |
Number of A/C/T/G bases in the annotated sequence | |
mean_map |
Mean mappability for the annotated feature. Calculated from the configured mappability bedgraph file | |
tx_rnk |
Rank in the transcript | FX+SJ only |
num_exons |
Number of exons in tx; 1,2,3,4,5,>5 | |
tx_mappability |
Transcript mappability, factor with levels: low, medium, high | FX+SJ only |
len |
Length of annotated feature | |
mappability |
Annotation mappability, factor with levels: low, medium, high | |
GC |
Fraction of G/C for annotated feature | |
frac_convertible |
Fraction of convertible bases for annotation | SJ only |
convertibility |
Convertibility, factor with levels: low, medium, high | |
don_ex_A/C/T/G |
Number of A/C/T/G bases in exonic part of donor window (genomic window centred on splice donor site with size: 2xreadlen+1) |
SJ only |
don_in_A/C/T/G |
Number of A/C/T/G bases in intronic part of donor window | SJ only |
don_win_map |
Mean mappability of donor window | SJ only |
don_mappability |
Donor window mappability, factor w levels: low, medium, high | SJ only |
don_ex_fc |
Fraction of convertible bases in the exonic part of the donor window | SJ only |
don_in_fc |
Fraction of convertible bases in the intronic part of the donor window | SJ only |
ac_* |
Analogous to the splice donor columns above, but for splice acceptor site | SJ only |
Splice_sim simulates nucleotide conversions in reads based on Bernoulli processes with given (configured) conversion probabilities.
While we believe this to be appropriate for simulating BS-seq or SLAM-seq data (see our paper), it might not be suitable for other use-cases where, e.g., conversion prob abilities are affected by local sequence context (e.g., RNA-editing) or the like.
For customization of the NC simulation process, users may alter/extend the splice_sim.simulator.modify_bases method that has access to
- the sequence of the simulated read (w/o NC)
- genomic coordinates and orientation (strand) of the simulated read
- configured NC reference and alternate bases
- the configured conversion_rate
- a list of convertible positions in the read
- a list of SNPs that affect this read (if configured)