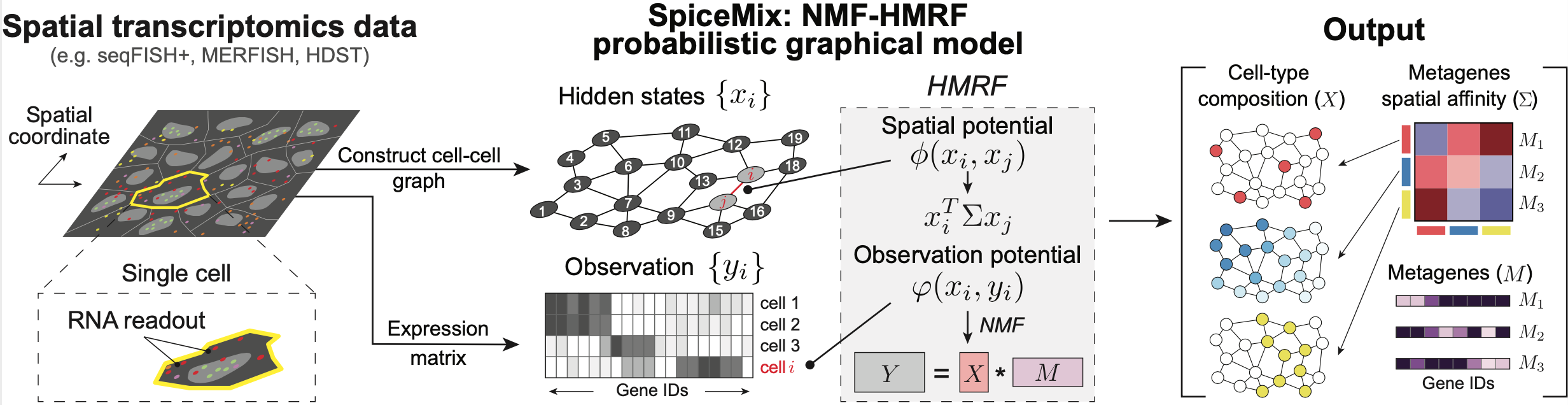
SpiceMix
SpiceMix is an unsupervised tool for analyzing data of the spatial transcriptome. SpiceMix models the observed expression of genes within a cell as a mixture of latent factors. These factors are assumed to have some spatial affinity between neighboring cells. The factors and affinities are not known a priori, but are learned by SpiceMix directly from the data, by an alternating optimization method that seeks to maximize their posterior probability given the observed gene expression. In this way, SpiceMix learns a more expressive representation of the identity of cells from their spatial transcriptome data than other available methods.
SpiceMix can be applied to any type spatial transcriptomics data, including MERFISH, seqFISH, and HDST.
Requirement
- Python=3.7.3
- scipy=1.2.1
- psutil=5.6.3
- gurobi=8.1.1
- pytorch=1.0.1
- numpy=1.16.2
- scikit-learn=0.21.1
Running SpiceMix
All code is contained within the SpiceMix folder. The script main.py runs the program -- see details for example usage.
Step 1: preparing input files
All files for one run of SpiceMix must be put into one directory. For each FOV with name <FOV> of N cells and G genes, the following two files stored as tab-delimited txt format are required for SpiceMix:
expression_<FOV>_<expr_suffix>.txt, an N-by-G nonnegative-valued matrix of normalized single-cell expression profiles. In our paper, we applied the following steps of normalization to all data sets:neighborhood_<FOV>_<neigh_suffix>.txt, a neighbor graph represented as a list of cell pairs, i.e., an|E|-by-2 integer-valued matrix. Cells are assigned with integer indices starting from 0 in the order that they appear in the expression profile file. We recommend the following two methods to generate the neighbor graph from cells' spatial coordinates:- K-nearest neighbor graph under Euclidean metric
- Delaunay triangulation followed by discarding interactions between cells that are far away from each other
When there are multiple isolated FOVs, the order of genes in expression_<FOV>_<expr_suffix>.txt for all FOVs must be identical.
The FOV name, denoted by <FOV> here, is required in order to distinguish between cells from different FOVs, especially when they don't share the same coordinate system. FOV names can be any string, including '1', 'cortex', 'mouse brain Oct-10-2020'.
To compare different normalizations and neighbor graphs, the two suffices, <expr_suffix> and <neigh_suffix>, can be used to distinguish between normalizations and neighbor graphs, respectively. For example, neighborhood_3_KNN.txt and neighborhood_3_Delaunay.txt may denote the neighbor graph generated by k-nearest neighbor and Delaunay triangulation, respectively.
The two suffices are optional, and the preceding underscore _ should be absent when the corresponding suffix is an empty string.
The following data are used in downstream analysis and visualization:
- Gene names or symbols. These can be stored in a multi-line file with one gene per line. The order of genes should match that in
expression_<FOV>_<expr_suffix>.txt. - Cells' spatial coordinates in the 2D or 3D space.
Step 2: Organizing files
In the root directory, two directories should be created and named scripts and data. In the data directory, every data set of one or multiple FOVs should have its own directory, named by the name of the data set. In the directory of each data set, two directories, named files and logs, need to be created. The files directory contains input files prepared in step 1, and the log directory is initially empty.
For example, when there are two data sets, named simulation 1 and mouse primary visual cortex, consisting of 3 and 2 FOVs, respectively, the file structure of Python scripts and input files should be as following:
.
├── scripts
│ ├── main.py
│ ├── readData.py
│ └── ...
└── data
├── simulation 1
| ├── files
| | ├── expression_1.txt
| | ├── neighborhood_1.txt
| | ├── expression_2.txt
| | ├── neighborhood_2.txt
| | ├── expression_3.txt
| | └── neighborhood_3.txt
| └── logs
└── mouse primary visual cortex
├── files
| ├── expression_A_allgenes.txt
| ├── expression_A_top100genes.txt
| ├── neighborhood_A_KNN.txt
| ├── neighborhood_A_Delaunay.txt
| ├── expression_B_allgenes.txt
| ├── expression_B_top100genes.txt
| ├── neighborhood_B_KNN.txt
| ├── neighborhood_B_Delaunay.txt
└── logs
Step 3: Inferring latent states, metagenes, and pairwise affinity matrix
SpiceMix requires a few arguments that specify input files and hyperparameters. Below is the table of description of all arguments:
Input related parameters
| params | type | description | example |
|---|---|---|---|
| --dataset | str | name of the dataset | "simulation 1" or "mouse primary visual cortex" |
| --neighbor_suffix | str | suffix of the name of the file that contains interacting cell pairs | "", "KNN", or "Delaunay" |
| --expression_suffix | str | suffix of the name of the file that contains expressions | "", "allgenes", or "top100genes" |
| --exper_list | list of strings (Python expression) | A Python expression of a list of FOV names | "[0,1,2]", "[A,B]", or "range(3)" |
| --use_spatial | list of booleans (Python expression) | A Python expression of a list of boolean variables controlling whether to use the neighbor graph for each FOV | "[True,True,True]", "[False]*5", or "[True,False,True]" |
Hyperparameters
| params | type | description | example |
|---|---|---|---|
| -K | int | dimension of latent space | 20 |
| --lambda_SigmaXInv | float | regularization on Sigma_x^{-1} | 1e-4 |
| --max_iter | int | maximum number of coordinate optimization iterations | 200 or 1000 |
| --init_NMF_iter | int | number of NMF iterations times 2: an odd number will include an extra optimation of latent states | 5 |
| --beta | list of floats (Python expression) | A Python expression of a list of real-values weights for FOVs, the weight will be normalized to be sum-to-one | "[1,1,1]", "[1,10]" |
Reproducibility related parameters
| params | type | description | example |
|---|---|---|---|
| --random_seed4kmeans | int | the initial random seed fed to k-means | 0 |
Output control
| params | type | description | example |
|---|---|---|---|
| --logger | none | to save inferred parameters into files rather than outputing them to screen | N/A |
To run SpiceMix, use the following command filled with proper value for each argument in the script directory, e.g.,
python SpiceMix.py -K=20 --dataset="simulation 1" --exper_list="[1,2]" --use_spatial="[True]*2" --logger
Locating results
The output of every SpiceMix run is saved into a directory, named by date, time, and PID, in the logs directory. After the ith iteration of the coordinate descent and the initialization (denoted by iteration i=0), three pickle files are produced:
Q_<i>.pkl, which contains a single real number which is the negative logarithm of the joint probabilityH_<i>.pkl, which contains inferred latent states. In this file is tuple whose only element is a list of Numpy matrices, one for each FOV. For theith FOV ofNicells, theith matrix in the list has dimensionNi-by-K. The cells appear in the same order as inexpression_<FOV>_<expr_suffix>.txt.Theta_<i>.pkl, which contains model parameters stored in a tuple of 5 elements. The 5 elements are:- Metagenes, a
G-by-Knonnegative matrix such that each column sums to one. - Noise levels
σ_yx^-1, an array of length equal to the number of FOVs. Currently, the noise levels of all FOVs are assumed to be the same. - Pairwise affinity matrix
Σ_x^-1, aK-by-Ksymmetric matrix, where negative values indicate appealing. - Placeholder of an obsolete parameter.
- Prior of latent. Currently, all dimensions of the latent space are assumed to have exponential distribution with parameter 1.
- Metagenes, a
Cite
Cite our paper by
@article{chidester2020spicemix,
title={SPICEMIX: Integrative single-cell spatial modeling for inferring cell identity},
author={Chidester, Benjamin and Zhou, Tianming and Ma, Jian},
journal={bioRxiv},
year={2020},
publisher={Cold Spring Harbor Laboratory}
}