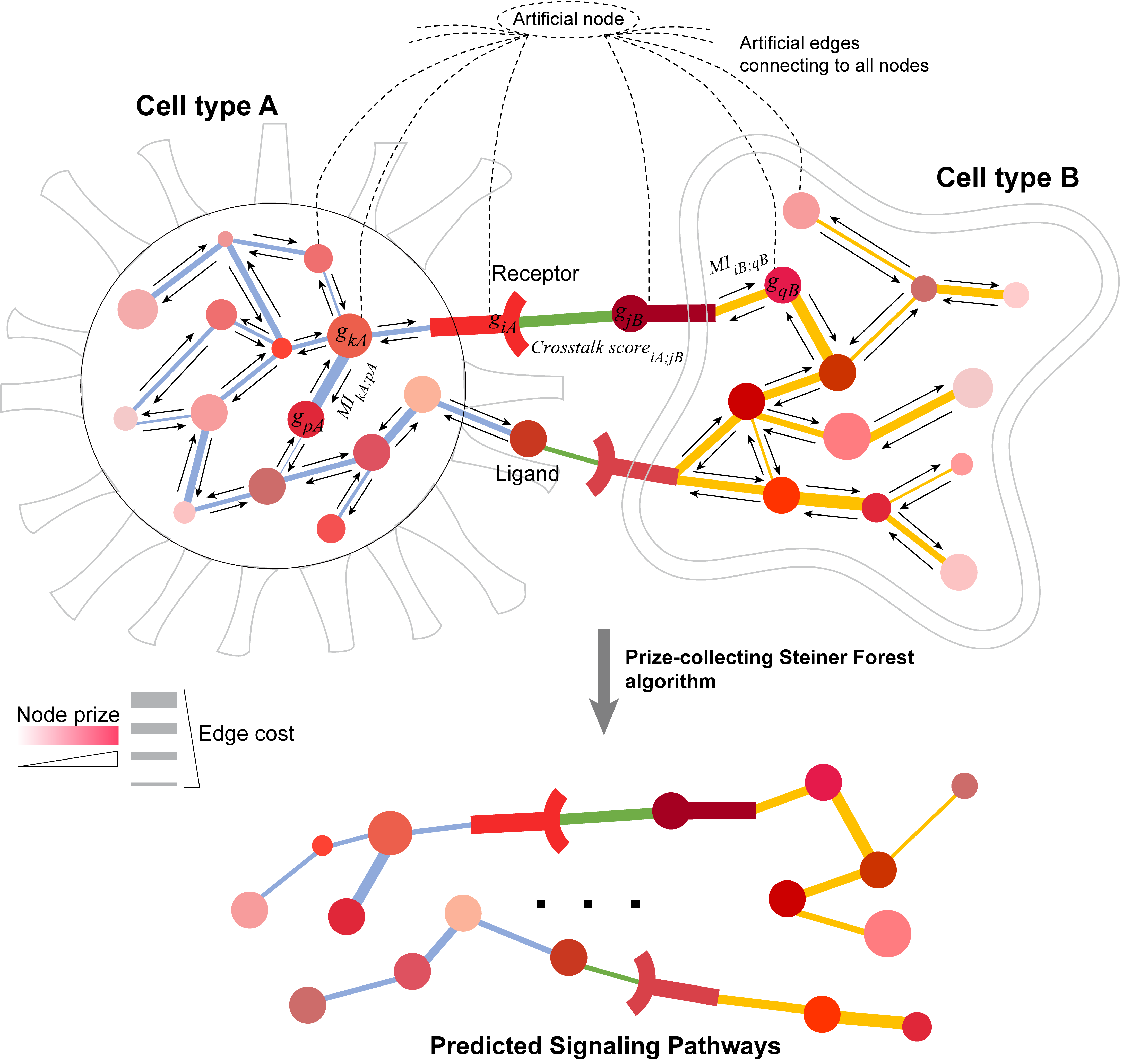
We have developed the CytoTalk algorithm for de novo construction of a signaling network (union of multiple signaling pathways emanating from the ligand- receptor pairs) between two cell types using single-cell transcriptomics data. The algorithm constructs an integrated network of intracellular and intercellular functional gene interactions. The signaling network is identified by solving a prize-collecting Steiner forest (PCSF) problem based on appropriately defined node prize (i.e. cell-specific gene activity) and edge cost (i.e. functional interaction between two genes). The objective of the PCSF problem is to find an optimal subnetwork in the integrated network that includes genes with high levels of cell-type-specific expression and close connection to highly active ligand-receptor pairs.
Signal transduction is the primary mechanism for cell-cell communication. scRNA- seq technology holds great promise for studying cell-cell communication at much higher resolution. Signaling pathways are highly dynamic and cross-talk among them is prevalent. Due to these two features, simply examining expression levels of ligand and receptor genes cannot reliably capture the overall activities of signaling pathways and interactions among them.
CytoTalk requires a Python module to operate correctly. To install the
pcst_fast module, please
run this command before using CytoTalk:
pip install git+https://github.com/fraenkel-lab/pcst_fast.gitCytoTalk outputs a SIF file for use in Cytoscape. Please install
Cytoscape to view the whole output
network. Additionally, if you want the final ligand-receptor pathways to
render portable SVG files correctly, you’ll have to have Graphviz
installed and the dot executable on your PATH. See the Cytoscape
downloads page for more information.
If you have devtools installed, you can use the install_github
function directly on this repository:
devtools::install_github("tanlabcode/CytoTalk", build_vignettes = TRUE)Let’s assume we have a folder called “scRNAseq-data”, filled with single-cell RNA sequencing (scRNASeq) datasets. Here’s an example directory structure:
── scRNAseq-data
├─ scRNAseq_BasalCells.csv
├─ scRNAseq_BCells.csv
├─ scRNAseq_EndothelialCells.csv
├─ scRNAseq_Fibroblasts.csv
├─ scRNAseq_LuminalEpithelialCells.csv
├─ scRNAseq_Macrophages.csv
└─ scRNAseq_TCells.csv⚠ IMPORTANT ⚠
Notice all of these files have the prefix “scRNAseq_” and the extension “.csv”; CytoTalk looks for files matching this pattern, so be sure to replicate it with your filenames. If you want to make sure your names are valid, use the following function:
dir_in <- "~/scRNAseq-data"
CytoTalk::check_valid_names(dir_in)
#> [1] "BasalCells" "BCells" "EndothelialCells"
#> [4] "Fibroblasts" "LuminalEpithelialCells" "Macrophages"
#> [7] "TCells"The outputted names are all the cell types we can choose to run CytoTalk against. We’ll come back to this.
⚠ IMPORTANT ⚠
Each of these datasets should have the same number of rows, the first column corresponds to gene names, and the gene names of all these expression matrices should be exactly the same. Additionally, the gene expression values should be ln-transformed, normalized scRNA-seq data (e.g. Seurat-preprocessed data).
Here is a small excerpt of the “scRNAseq_BCells.csv”, for reference:
, 10X_P7_12_AAACCTGTCGTCACGG,10X_P7_12_AAACGGGCAGTGGGAT
Xkr4, 0, 0
Rp1, 0, 0
Sox17, 0, 0
Mrpl15,0.71676, 1.29024
Lypla1,0, 0
Tcea1, 0, 0
Notice the first column has no header, this is fine.
Without further ado, let’s run CytoTalk!
# set required parameters
dir_in <- "~/scRNAseq-data"
type_a <- "BCells"
type_b <- "TCells"
# run CytoTalk process
CytoTalk::run_cytotalk(type_a, type_b, dir_in)We selected cell types “BCells” and “TCells”, specified our input
directory, and that’s all we need for a default run (recommended
settings). CytoTalk will automatically create a new folder called
“CytoTalk-output” in our working directory, but we can set the dir_out
parameter to change this behavior. The most important optional
parameters to look at are cutoff_a, cutoff_b, and beta_max;
details on these can be found in the help page for the run_cytotalk
function.
As the process runs, we see messages print to the console for each sub process. At the same time, we can view our output directory in a file explorer and watch as it propagates with files. The process can be stopped at any time, and it will start where it left off. If we would like files re-computed, we can either delete them or specify a new output directory.
Here is what the resulting output directory structure looks like (abbreviated):
── CytoTalk-output
├─ cytoscape
│ ├─ CytoscapeEdges.txt
│ ├─ CytoscapeNetwork.sif
│ └─ CytoscapeNodes.txt
├─ graphviz
│ ├─ CD48_CD2.gv
│ ├─ CD48_CD2.png
│ ├─ CD48_CD2.svg
│ └─ ...
├─ IntegratedNetwork.cfg
├─ PCSF_Network.txt
└─ ...In the order of increasing effort, let’s take a look at some of the
results. Let’s begin with the “graphviz” subfolder. This folder contains
source GV files, which the dot executable has also transformed into
PNG images and SVG vector graphics (also includes hyperlink support).
Here is an example pathway neighborhood:
Note that the SVG files are interactive, with hyperlinks to GeneCards and WikiPI. Green edges are directed from ligand to receptor. This is a subset of the overall network, so how do we view the whole thing?
We have the “cytoscape” folder, which includes a SIF file read to import and two tables that can be attached to the network and used for styling. Here’s an example of a styled Cytoscape network:
There are a number of details we can glean from these graphs, such as node prize (side of each node), edge cost (inverse edge width), Preferential Expression Measure (intensity of each color), cell type (based on color, and shape in the Cytoscape output), and interaction type (dashed lines for crosstalk, solid for intracellular).
Finally, we can take a look at some of the textual output. Most of the text files found in the output folder are used for intermediate calculations, but they are provided in case you want to check our work. The final network (includes edges and node attributes) generated by the prize-collecting Steiner tree (PCST) algorithm can be found in the “PCSF_Network.txt” file. If you would like to see the inputs to the PCST algorithm, check out the “PCSF_Network.txt” file, but note that this does not include the artificial node that connects all others together.
2021-10-07: The latest release “CytoTalk_v4.0.0” is a completely re-written R version of the program. Approximately half of the run time as been shaved off, the program is now cross-compatible with Windows and *NIX systems, the file space usage is down to roughly a tenth of what it was, and graphical outputs have been made easier to import or now produce portable SVG files with embedded hyperlinks.
2021-06-08: The release “CytoTalk_v3.1.0” is a major updated R version on the basis of v3.0.3. We have added a function to generate Cytoscape files for visualization of each ligand-receptor-associated pathway extracted from the predicted signaling network between the two given cell types. For each predicted ligand-receptor pair, its associated pathway is defined as the user-specified order of the neighborhood of the ligand and receptor in the two cell types.
2021-05-31: The release “CytoTalk_v3.0.3” is a revised R version on the basis of v3.0.2. A bug has been fixed in this version to avoid errors occurred in some special cases. We also provided a new example “RunCytoTalk_Example_StepByStep.R” to run the CytoTalk algorithm in a step-by-step fashion. Please download “CytoTalk_package_v3.0.3.zip” from the Releases page (https://github.com/huBioinfo/CytoTalk/releases/tag/v3.0.3) and refer to the user manual inside the package.
2021-05-19: The release “CytoTalk_v3.0.2” is a revised R version on the basis of v3.0.1. A bug has been fixed in this version to avoid running errors in some extreme cases. Final prediction results will be the same as v3.0.1. Please download the package from the Releases page (https://github.com/huBioinfo/CytoTalk/releases/tag/v3.0.2) and refer to the user manual inside the package.
2021-05-12: The release “CytoTalk_v3.0.1” is an R version, which is more easily and friendly to use!! Please download the package from the Releases page (https://github.com/huBioinfo/CytoTalk/releases/tag/v3.0.1) and refer to the user manual inside the package.
-
Hu Y, Peng T, Gao L, Tan K. CytoTalk: De novo construction of signal transduction networks using single-cell transcriptomic data. Science Advances, 2021, 7(16): eabf1356.
-
Hu Y, Peng T, Gao L, Tan K. CytoTalk: De novo construction of signal transduction networks using single-cell RNA-Seq data. bioRxiv, 2020.
- Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research, 2003, 13: 2498-2504.
Kai Tan, tank1@chop.edu