Simulate fragment libraries (Illumina SE/PE/MP, Pacbio, contigs) from reference sequences. Errors models are not currently supported.
cpanm Math::Random
# wherever you prefer
git clone https://github.com/BioInf-Wuerzburg/perl5lib-Fasta.git
git clone https://github.com/BioInf-Wuerzburg/perl5lib-Fasta.git
export PERL5LIB=/path/to/perl5lib-Fasta/Fastq:$PERL5LIBseq-frag MODE -l LENGTH -c COVERAGE [options ..] < FASTA
# 50X 100bp single end reads
seq-frag se -l 100 -c 50 < genome.fa > read.fq
# 50X 100bp paired end, 180bp insert
seq-frag pe -l 100 -c 50 -i 180 < genome.fa | interleaved-split 1>reads_1.fq 2>reads_2.fq
# 50X mate pair, 2000bp insert
seq-frag mp -l 100 -c 50 -i 2000 < genome.fa | interleaved-split 1>reads_1.fq 2>reads_2.fq
# 20X pacbio style fragments, mean length 2000bp
seq-frag pacbio -l 2000 -c 20 < genome.fa > pb-reads.fq
# for more details
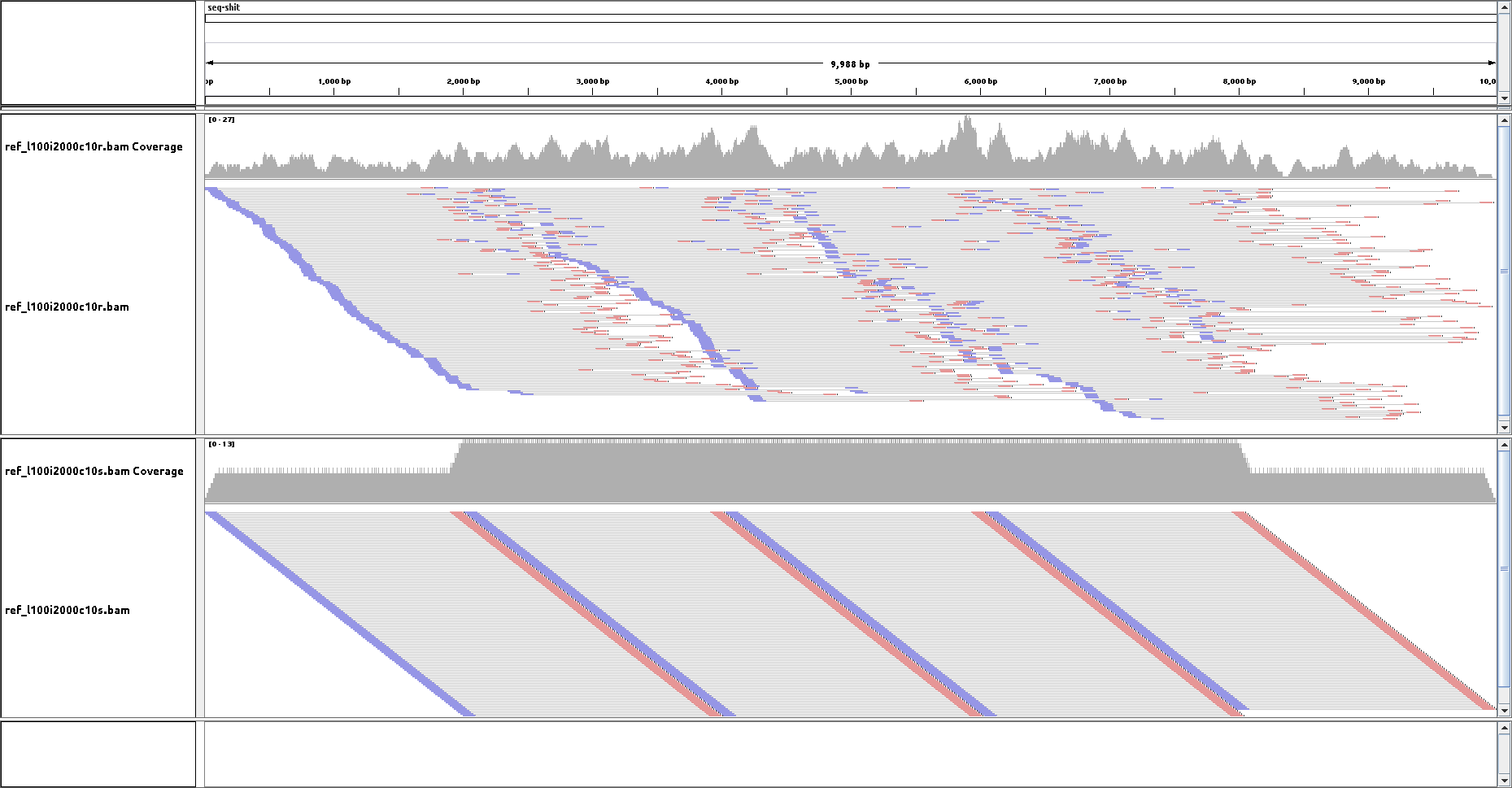
seq-frag --helpseq-frag mp -l 100 -c 1 -i 2000 ref.fa | interleaved-split 1> r_1.fq 2> r_2.fq
seq-frag mp -l 100 -c 1 -i 2000 -s ref.fa | interleaved-split 1> s_1.fq 2> s_2.fqMapped with bwa mem and visualized with IGV:
Plot mappings (bam), features and annotations (gff, bed) along sequences to high quality SVGs.
git clone https://github.com/thackl/perl5lib-Gff.git
git clone https://github.com/thackl/perl5lib-SVG-Bio.git
export PERL5LIB=/path/to/perl5lib-Gff/lib:/path/to/perl5lib-SVG-Bio/lib:$PERL5LIB;
bio2svg --fasta FA --region REGION --gff GFF --bam BAM > SVGRegion can be either a sequence ID (Chr4) or an ID with range (Chr4:521-15521).
bio2svg --width 10000 --fa MaV-is-CrEc-001.ctg.fa --region MaV-is-CrEc-001 \ --gff mav-regions.gff --gff MaV-gen-2.0.maker.lifted.gff \ --gff CrEc-gen-dp-1.0.all.lifted.gff \ --bam PCR\~MaV-is-CrEc-001.ctg.bam \ --bam pr-all\~MaV-is-CrEc-001.ctg-support.bam \ --bam pr-all~MaV-is-CrEc-001.ctg-bridge.bam \ > MaV-is-CrEc-001.ctg.svg