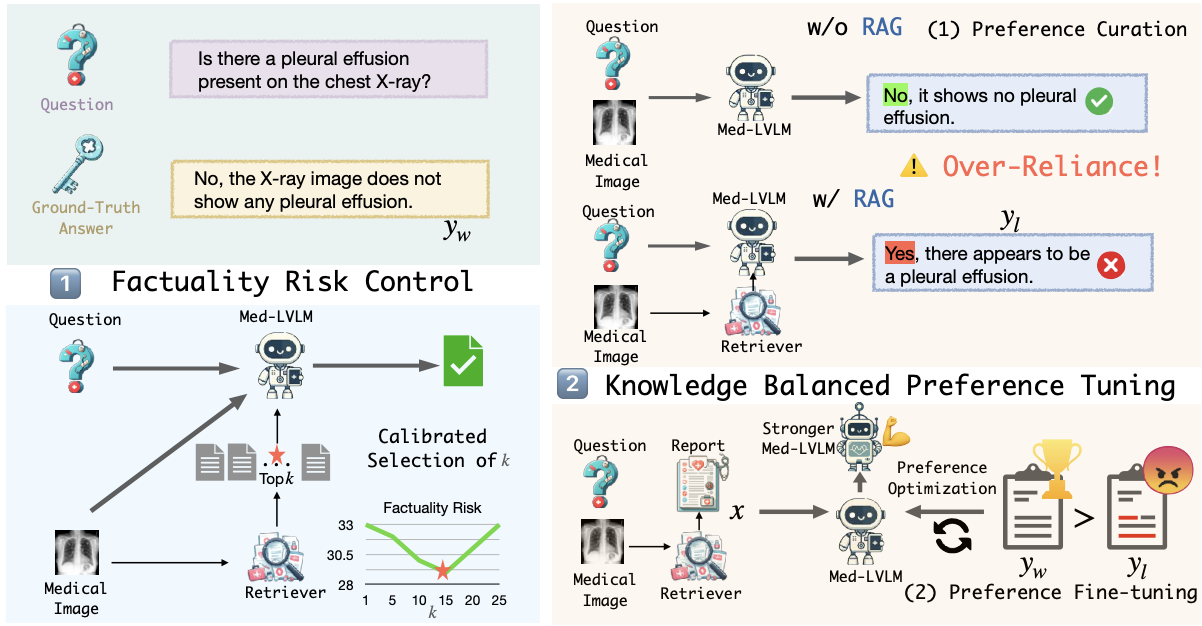
We tackle the challenge of improving factual accuracy in Medical Large Vision Language Models (Med-LVLMs) using our novel approach, RULE. Despite their promise, Med-LVLMs often generate responses misaligned with established medical facts. RULE addresses this with two key strategies: 1) Calibrated selection of retrieved contexts to control factuality risk. 2) Fine-tuning models using a preference dataset to balance reliance on inherent knowledge and retrieved contexts. Our method achieves a 20.8% improvement in factual accuracy across three medical VQA datasets.
RULE: Reliable Multimodal RAG for Factuality in Medical Vision Language Models [Paper]
- Clone this repository and navigate to RULE folder
git clone https://github.com/richard-peng-xia/RULE.git
cd RULE- Install Package: Create conda environment
conda create -n RULE python=3.10 -y
conda activate RULE
pip install --upgrade pip # enable PEP 660 support
pip install -e .
pip install trl- Download the model checkpoints LLaVA-Med-1.5 from huggingface.
- Download the test data and annotations under
data/. - Download the model checkpoints after DPO training under
checkpoints/.
- The training code of Direct Preference Optimization is at
llava/train/train_dpo.py. - The relevant script can be found at
scripts/run_dpo.sh
- For test dataset inference, you need to specify the following arguments.
python llava/eval/model_vqa_{dataset}.py \
--model-base 'path/to/llava-med-1.5' \
--model-path 'path/to/lora_weights' \
--question-file 'path/to/question_file.json' \
--image-folder 'path/to/test_images' \
--answers-file 'path/to/output_file.json'- The written script is at
scripts/inference.sh. Before that, you need to set the correct path of data and checkpoints in your script.
@article{xia2024rule,
title={RULE: Reliable Multimodal RAG for Factuality in Medical Vision Language Models},
author={Xia, Peng and Zhu, Kangyu and Li, Haoran and Zhu, Hongtu and Li, Yun and Li, Gang and Zhang, Linjun and Yao, Huaxiu},
journal={arXiv preprint arXiv:2407.05131},
year={2024}
}We use code from LLaVA-Med, POVID, CARES. We thank the authors for releasing their code.