Phantom is a simple python module intended to simplify medical DICOM anonymization for medical machine-learning applications. Please keep in mind that we do NOT guarantee IRB-validated outputs and are not liable for any breaches of patient privacy.
git clone https://github.com/cyrilzakka/Phantom.git
cd PhantomPhantom relies on a few dependencies to run:
pip install -r requirements.txtPhantom maintains a very strict paradigm for anonymization to ensure a maximum degree of privacy. Identifiers specified in a base.yaml are kept within the metadata of the DICOM file, while everything else, inluding private metadata is purged. Pixel data is always preserved.
To anonymize DICOM metadata, simply run:
python meta/metadata.py -s /path/to/dicoms -c /path/to/config.yamlwhere config.yaml is a simple YAML specifying the attributes to keep in the DICOM.
--- base.yaml ---
base:
keep: ['PatientsSex'] # keys to keep in the DICOM metadata list<str>
jitter: {'PatientBirthDate': 30} # keys to jitter specified as a dict dict<str:int>
generate: ['PatientID'] # keys to replace with a randomized ID list<str>As DICOMs become more complex, we resort to modularity to keep anonymization manageable. Phantom configs can be composed together to create more complex anonymization pipelines. As an example, here we create a new echo.yaml configuration that inherits from base.yaml above. while appending modality-specific attributes like HeartRate or NumberOfFrames. Please keep in mind that in the case of inheritance any keys specified within either configuration files will be kept.
--- echo.yaml ---
echo:
defaults: 'base'
keep: ['HeartRate', 'NumberOfFrames']
jitter: {'AcquisitionDateTime': 30, 'StudyDate': 30}
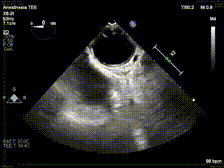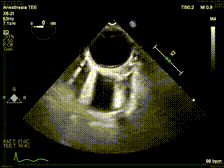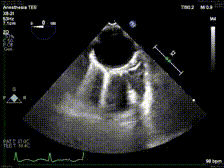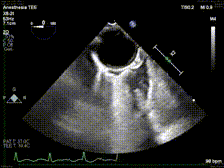
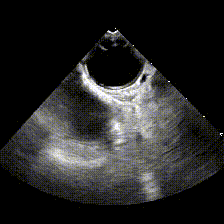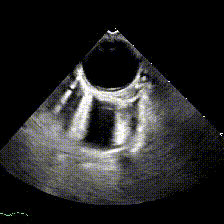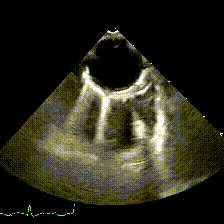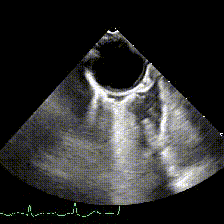
generate: [] DICOM modalities occasionally contain burned annotations, or patient data embedded within the pixels of the images. While this is often difficult to detect and remove without some sort of machine-learning approach, medical imaging modalities with a temporal dimension (e.g. echocardiograms) offer a simple solution via static pixel masking. This can be quickly achieved using the TemporalMaskingPipeline:
python pixel/temporal_mask.py -s /path/to/dicoms -d /path/to/destinationThis project is still in the alpha phase of development and is likely to experience some breaking changes as a result. If you run into any errors, please make sure to update the package first before opening an issue.
If you have an issues, feature requests, or simply want to contribute, please do not hesitate to submit a pull request.