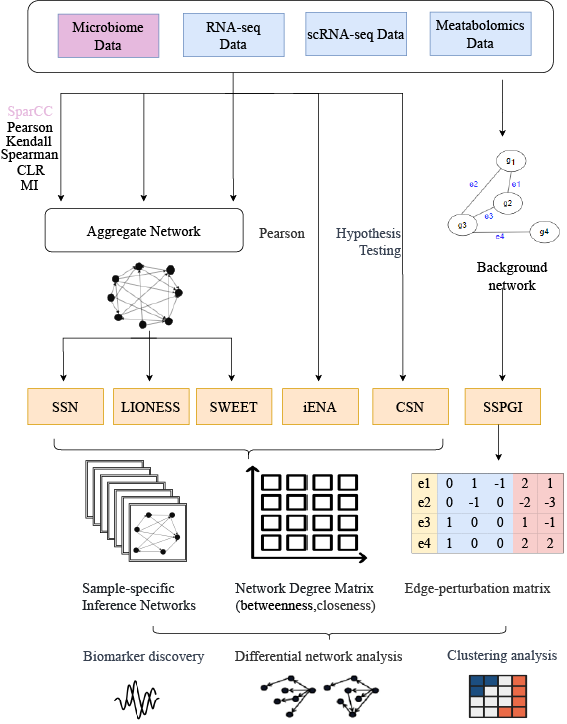
inteSIN: integrate sample-specific inference network methods. This package now includes SSN, LIONESS, SSPGI, iENA, CSN and Sweet. The flowchart is below.
- Huahui Ren
- Mingyue Zhao
This package can be installed using devtools.
devtools::install_github('rusher321/inteSIN')We welcome comments, criticisms, and especially contributions! GitHub issues are the preferred way to report bugs, ask questions, or request new features. You can submit issues here:
https://github.com/rusher321/inteSIN/issues
-
DNB function
-
network visualization
-
degree matrix visualization
-
other SIN methods test on microbiome datasets (CRC; Antibiotic; Infant...)
- collect 3 CRC cohorts/1 antibiotic/ 1 longitudinal infant (bacteria or virome) cohort and preprocess.
- the metaphlan3 profiling (XXX et al., XXX et al., ...) from curatedMetagenomicData.
- 🦊low priority:the virome profiling of infants could be generated using Phanta , the data is in here.
- comparison between biomarkers based on expression data and degree data.
- performance of classification, one option is Stabl, which could integrate the degree matrix and expression matrix using the data fusion.
- antibiotic: Which specific sample networks have higher antibiotic resistance
- infant: the change of specific sample network during the early life
- A gut microbial signature for combination immune checkpoint blockade across cancer types
- collect 3 CRC cohorts/1 antibiotic/ 1 longitudinal infant (bacteria or virome) cohort and preprocess.
- Please report any issues or bugs.
- License: MIT
- Get citation information for
inteSINin R doingcitation(package = 'inteSIN') - Please note that this project is released with a Contributor Code of Conduct. By participating in this project you agree to abide by its terms.