Chen X, Wang Y, Sheng Y, Warren A, Gao S (2018). GPSit: An automated method for evolutionary analysis of nonculturable ciliated microeukaryotes. Molecular Ecology Resources. https://doi.org/10.1111/1755-0998.12750
https://github.com/seanchen607/GPSit
Contact: Xiao Chen (seanchen607@gmail.com)
Institute of Evolution & Marine Biodiversity, Ocean University of China, Qingdao 266003, CHINA
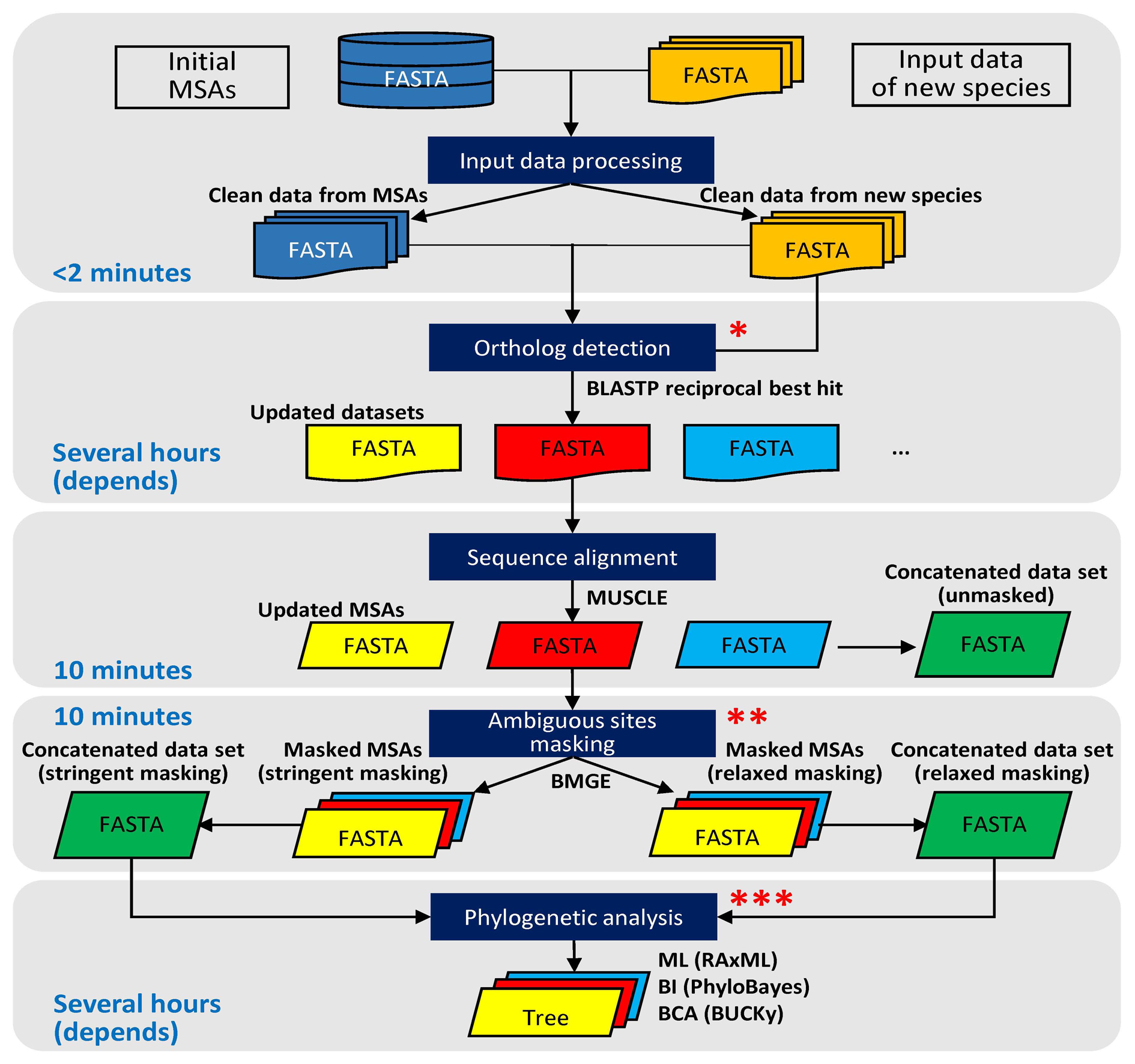
Guided Phylogenetic Search in tree (GPSit) is a simple and automated method to do phylogenomic reconstruction.
IMPORTANT: The GPSit method is implemented in Perl v5.16, please make sure the script is running under the Perl v5.16 environment.
perl GPSit.pl -i <input_dir1_initial_MSAs> -n <input_dir2_new_proteins> -o <output_dir> -b <BMGE_dir> [-optional parameters]
The arguments of GPSit are as follows:
-i <input_dir1_initial_MSAs>: [required] path to directory containing initial MSAs of original species (in FASTA format);
-n <input_dir2_new_proteins>: [required] path to directory containing protein data files of newly sequenced species (in FASTA format);
-o <output_dir>: [required] path to output directory;
-b <BMGE_dir>: [required] path to program "BMGE.jar" (BMGE v1.12);
-e <evalue_cutoff>: [optional] cutoff value of Evalue for BLASTP, default is 1e-10;
-d <ident_cutoff>: [optional] cutoff value of identity for BLASTP, default is 50;
-S: [optional] using stringent masking (gap rate cut-off = 30%, similarity matrix = BLOSUM62), instead of relaxed masking (default, gap rate cut-off = 50%, similarity matrix = BLOSUM30), when calling BMGE;
-M: [optional] jump to masking step (incompatible with option "-T");
-T: [optional] jump to tree-building step (incompatible with option "-M");
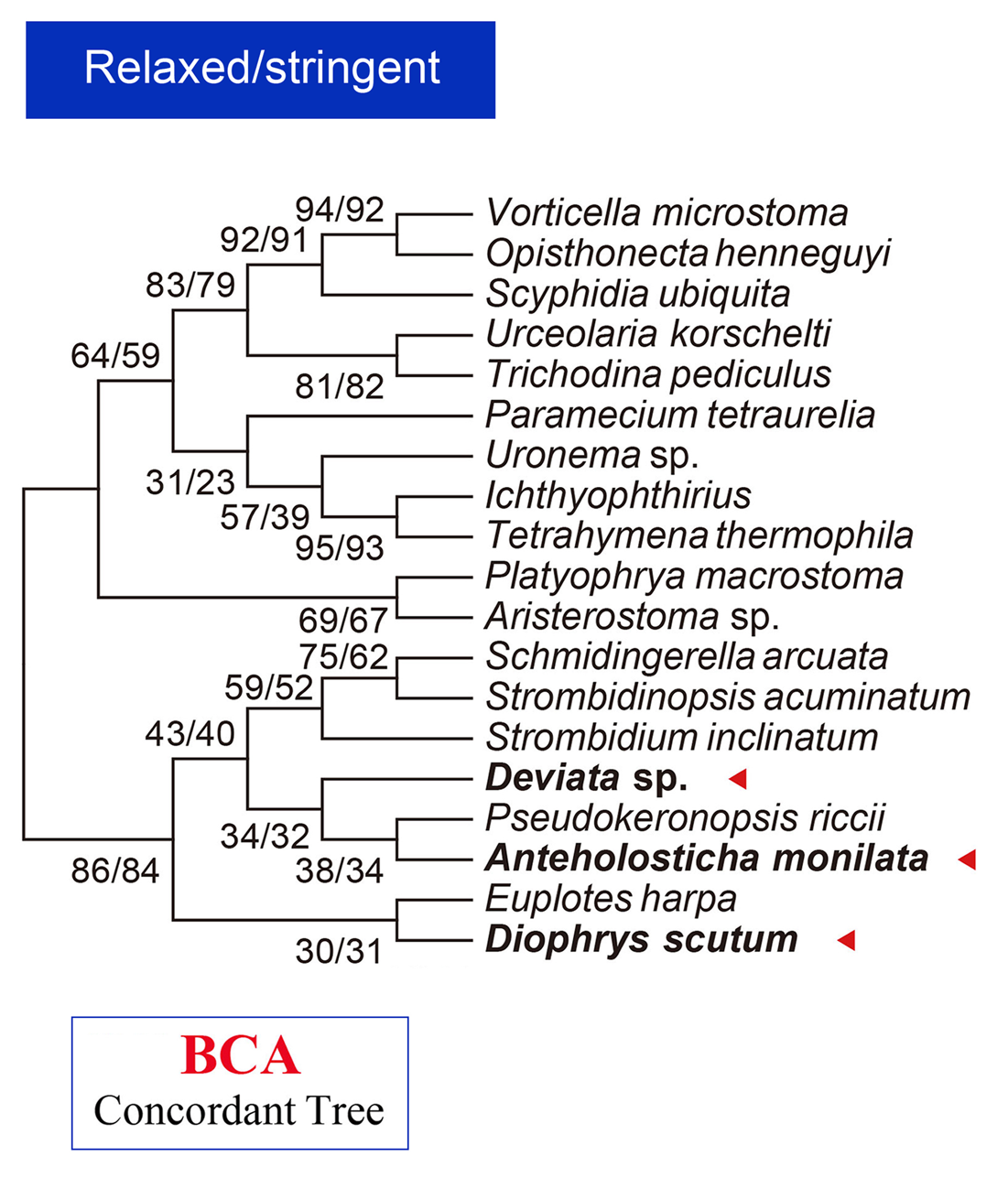
-U: [optional] call BUCKy to proceed Bayesian Concordance Analysis (BCA);
-l <taxa_list>: [optional, but required when "-U" is on] genes do not have all taxa in this specified list will be shipped in BCA analysis;
-g <generation_number>: [optional] integer, automatically enabled if "-U" is on, default is 100000;
-f <sample_frequency>: [optional] integer, automatically enabled if "-U" is on, default is 100;
-t <threads_numbr>: [optional] integer, default is 1.
Note: If the users are not using default parameters for -e/-d/-g/-f/-t, GPSit will use the default values. GPSit can be run simply as (not recommended):
perl GPSit.pl -i <input_dir1_initial_MSAs> -n <input_dir2_new_proteins> -o <output_dir> -b <BMGE_dir>
GPSit will call MUSCLE/MrBayes/BUCKy/BLASTP when running, please set up these seperate programs and set correct environment parameters before running GPSit.
MUSCLE==3.8.31
MrBayes==3.2.2
BUCKy==1.4.2
BLAST==2.3.0
# Environment settings before usage (please use practical paths of MUSCLE/MrBayes/BUCKy/BLASTP in the command lines below):
echo 'PATH=$PATH:{path_to_MUSCLE}:{path_to_MrBayes}:{path_to_BUCKy}:{path_to_BLASTP}' >> ~/.bashrc
echo 'LD_LIBRARY_PATH=/usr/local/lib:$LD_LIBRARY_PATH' >> ~/.bashrc
echo 'PKG_CONFIG_PATH=/usr/local/lib/pkgconfig:$PKG_CONFIG_PATH' >> ~/.bashrc
source ~/.bashrc
Imported MSA files of initial species and protein data files of newly sequenced species should be all in FASTA format, with ".fa" or ".fasta" as the suffixes of files.
Before we start, please make sure you have installed Perl v5.16 and use Linux as the operation system. GPSit should also work on Mac OS systems, but it is not tested.
Please download and uncompress the GPSit tarball and test.zip.
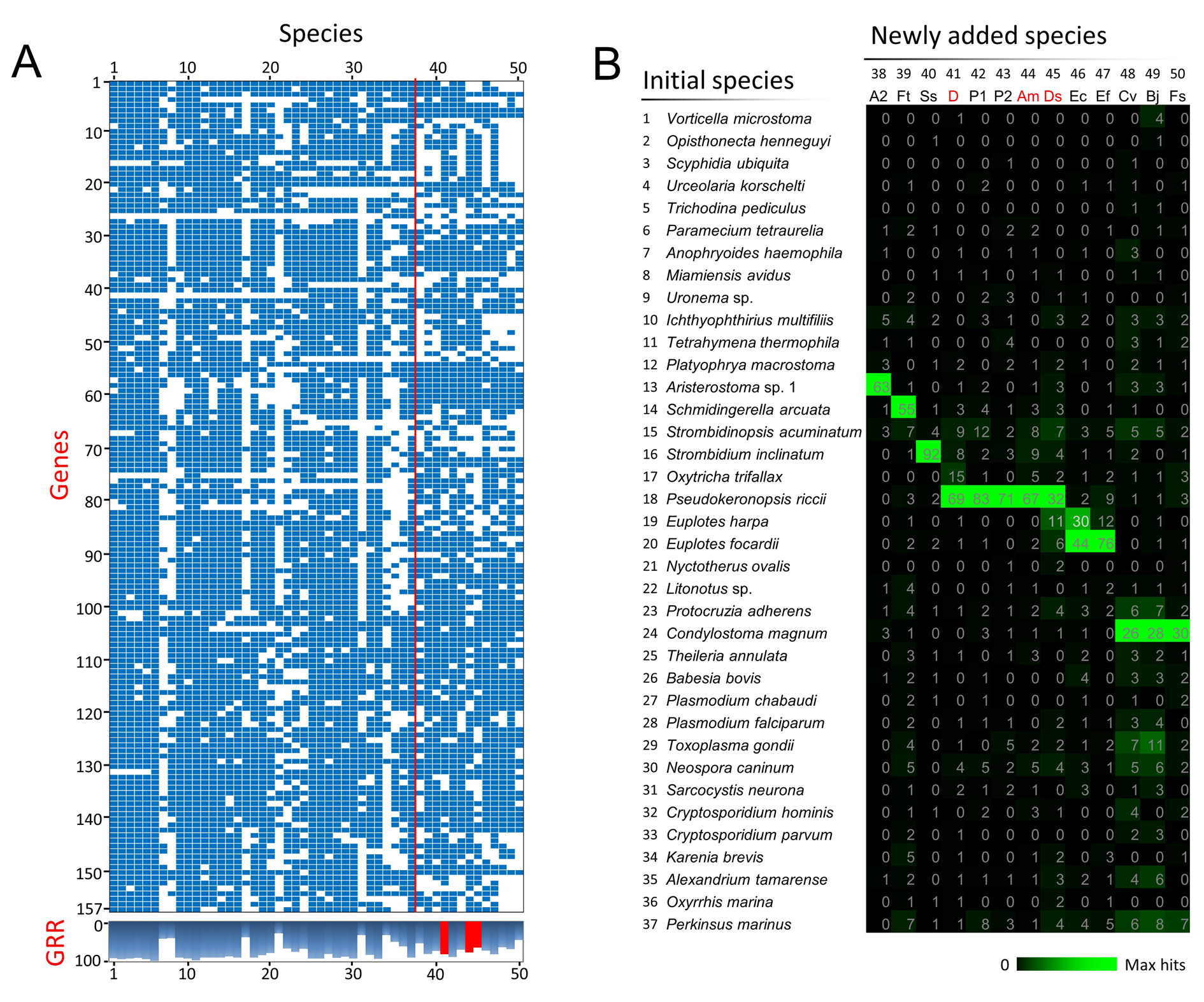
# The folder "test/01_initial_MSAs" contains 157 MSA files of 37 initial species.
# The folder "test/02_new_protein_seqs" contains 13 protein data files of newly sequenced species.
# The file "test/bucky_ciliates_generate80_3newsp.taxalist" contains 16 initial species whose gene recovery rates are above 80% and 3 newly sequenced species with single-cell sequencing data (for option "-l").
Enter the directory and type as following in your terminal:
# A new folder "03_output_dir" (as specified) will be created and output results will be deposited within it.
# Intermediate data files generated by GPSit are also recorded in the directory "${PATH_TO_GPSit}/test/03_output_dir/".
The command line can be as simple as:
perl GPStree.pl -i ${PATH_TO_GPSit}/test/01_initial_MSAs/ -n ${PATH_TO_GPSit}/test/02_new_protein_seqs/ -o ${PATH_TO_GPSit}/test/03_output_dir/ -b ${PATH_TO_BMGE}
# The folders and files would be automatically created in the directory "${PATH_TO_GPSit}/test/03_output_dir/".
00_log: contains all log files of GPSit.
01_clean_guide_prot: contains processed protein data of initial species.
02_clean_new_prot: contains processed protein data of newly sequenced species.
03_sh: contains shell scripts generated by GPSit.
04_blast: contains BLASTP results produced by GPSit.
05_unaligned: contains unaligned protein data for updating MSAs.
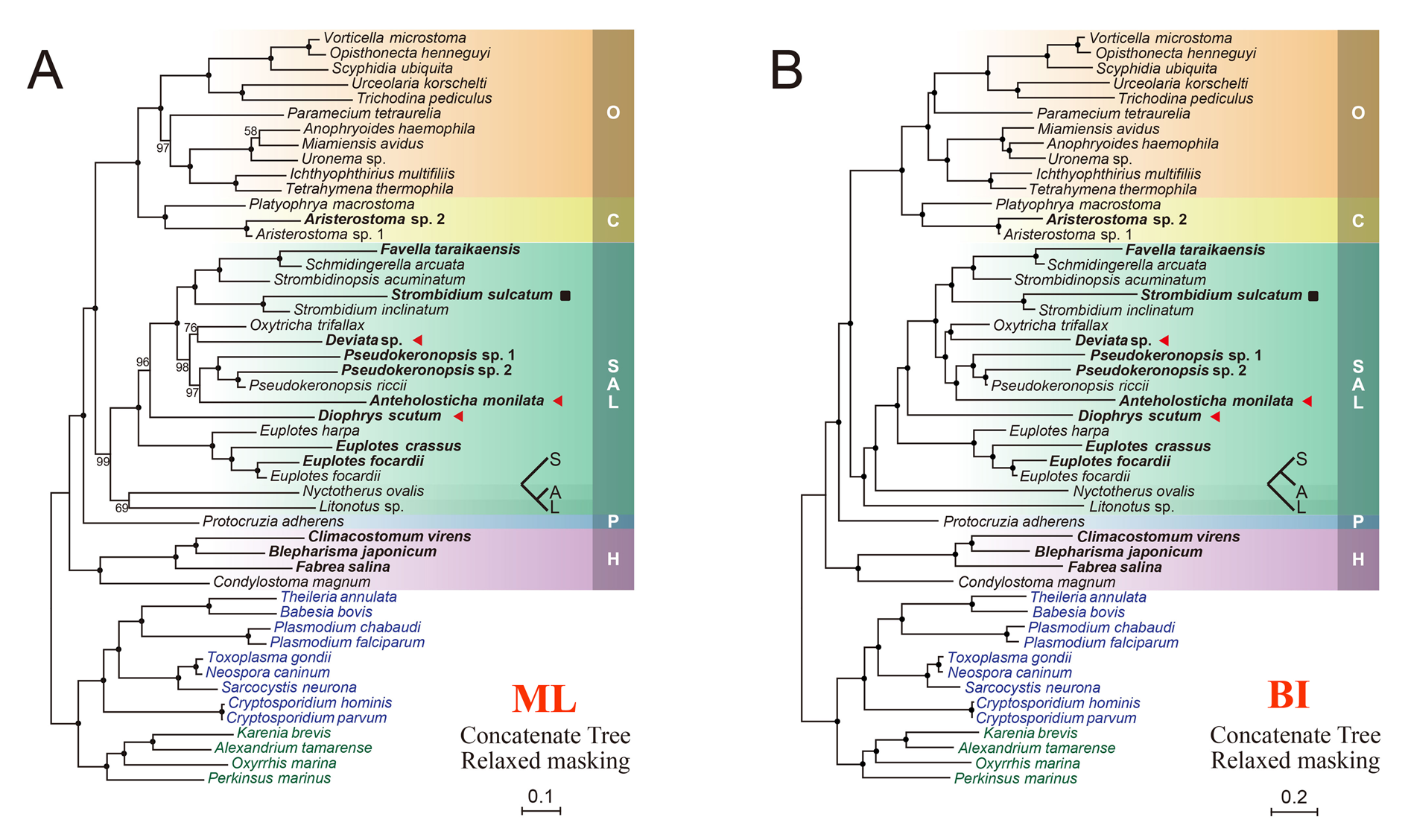
06_update_MSAs: [main output file] contains updated MSAs which integrated protein data of newly sequenced species (original), updated MSAs with ambiguous sites masked [for "supertree" methods, like Bayesian Concordance Analysis (BCA), implemented in BUCKy], as well as concatenated data sets after masking [for "supermatrix" methods, like maximum likelihood (ML) and Bayesian inference (BI), carried out by RAxML and PhyloBayes, respectively].
Update_MSAs_info.csv: contains the detailed information of updated MSAs.