Health Hack 2015
This is our server
ssh $USER@146.118.98.44
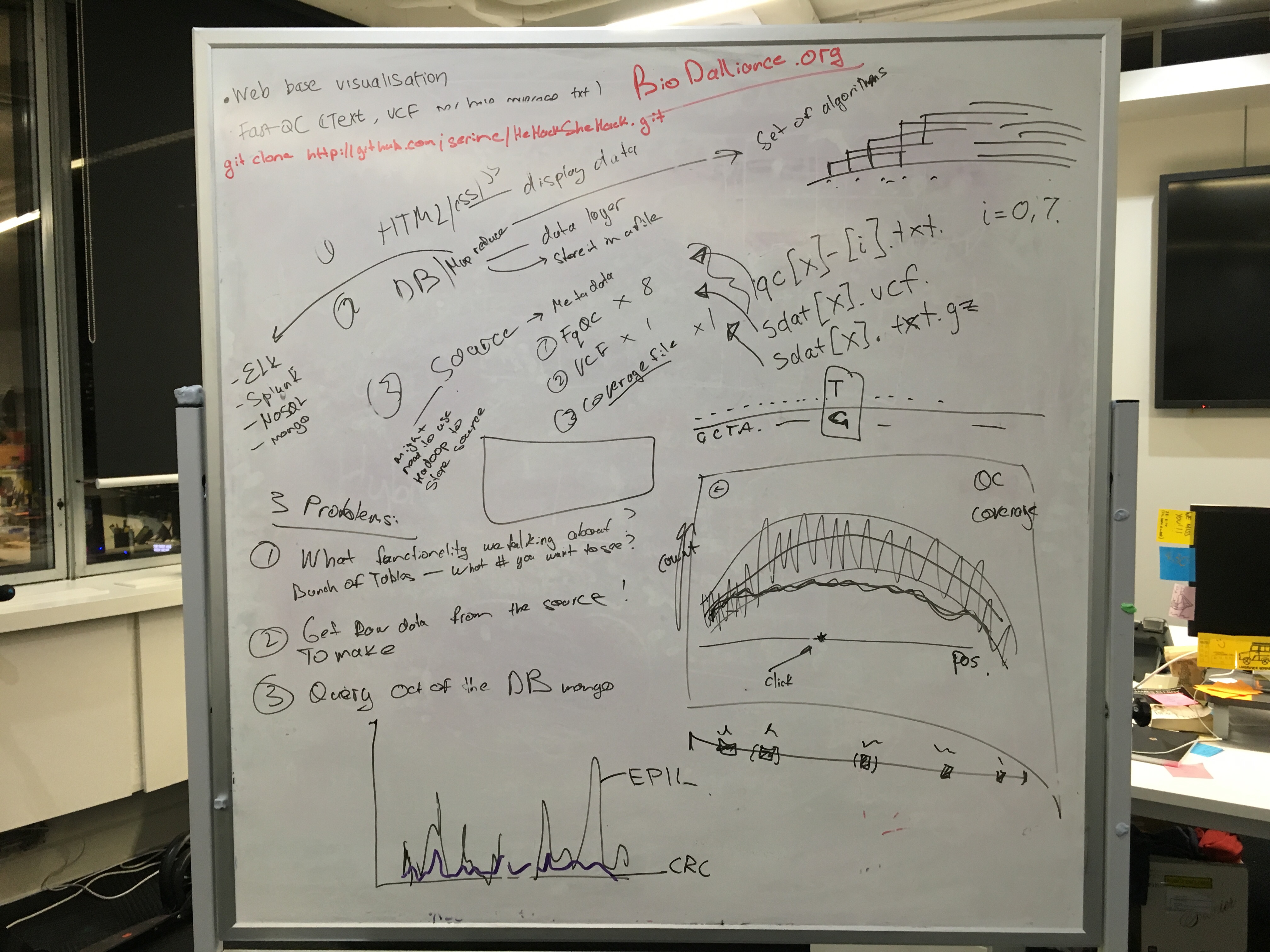
Our whiteborad plan
Aims:
- To write file parsers to parse fastqc, vcf and coverage files
- To write parsed output into mongoDB
- To query the database to extract the relevant info
- To visualise relevant information
Instructions for creating and using mongoDB
from pymongo improt MongoClient
connection = MongoClient("mongo://localhost:27017")
db_test = connection.healthhack.test
test_dict = {}
db_test.insert(test_dict)
results = db_test.find()
for result in results:
print resultlibrary(rmongodb)
query <- mongo.bson.from.list(list('city' = 'COLORADO CITY'))
mongo <- mongo.create()
mongo.get.databases(mongo)
mongo.get.database.collections(mongo, db)
db <- "healthhack"
res <- mongo.distinct(mongo, "healthhack.test_qc" , "uid")
test2 <- mongo.find.all(mongo, "healthhack.test_qc", list('uid' = "sdat89"))
pops1 <- mongo.find.all(mongo, "healthhack.test_qc", query = list('uid' = "sdat89", "quality" = list('$gte' = 32)))
test_qc <- mongo.find.all(mongo, "healthhack.test_qc")Git how to
- fork https://github.com/serine/HeHackSheHack this repo to your github
- git clone your version https://github.com/YOURUserName/HeHackSheHack.git
- don't delete any files just add your code files to that repo
git addyour filesgit commit -myour filesgit pushyour files to your own repo- send pull request
Hosting files from the server
If you like to host html or any other files put them into ~/www directory
and access it as such http://146.118.98.44/home/$USER/yourFile.html
General notes on the projects
- have individual track per patient
- show individual patient, but be able to take a subset of patients, for example ethnicity condition and/or other condition type
- make toggle box to see just a caucasian or asians
- have an average in the y-dimension may also consider average in the x-dimension
- have per gene colour block where you would show all possible variance, summarise all variance in that regions, for example have a heatmap cell, use heatmap colours
- bases shouldn't have colour - general notes
- if a block is more ered than in that gene (block) there are more mutations in that gene/block
- mutations burden - similar just a nubmer of mutation in that regions but also weighted
- base quality per experiment and is the least of our interest right now
Sticky notes notes
Take home message
two type of group
- pathologiest - don't care just want to see the result
- whereas developers they are interested what have you learn and technologies what are the pros and cons