An R package to locally serve copepod data from NOAA NMFS COPEPOD project. This package assists in the download of online datasets and simplifies local management.
Many of the data values are coded, so it is important the user has a working familiarity with the data source.
remotes::install_github("BigelowLab/copepoda")
The premise of this package is that data may be stored in a single
location but accessed by many users. To achieve this resource-friendly
goal and still simplified access for each user, we need to inform the
package where the data resides. We do this by storing the path to the
data location in each user’s home directory in a hidden text file,
“~/.copepoda”. That text file has just one line in it which contains
the full path to the shared dataset. For example, the author’s contains
/mnt/ecocast/coredata/noaa/nmfs/copepod which points to a shared
network drive mounted on our linux platform.
When the package is first loaded (ala library(copepoda)) the existence
of the file is check, and if missing a warning is issued.
You can create and populate that ~/.copepoda using a text editors, or
you can create using the provided function set_data_path(). Here is
how the author created his own…
library(copepoda)
copepoda::set_data_path("/mnt/ecocast/coredata/noaa/nmfs/copepod")
That’s it. If you ever move the data you’ll have to modify the contents of this hidden text file.
To fetch new, or update a local copy, you must know the dataset’s ID
(aka copepid). Below we fetch a dataset from the Scotian
Shelf
which matches copepid = ca-01003.
library(copepoda)
ok <- fetch_copepod(copepid = "ca-01003")Now the data directory will contain a subdirectory name
copepod__ca-01003. We can list the contents of the
short-form
dataset.
ff <- list_data(copepid = "ca-01003")
head(ff)## [1] "/mnt/ecocast/coredata/noaa/nmfs/copepod/copepod__ca-01003/data_src/short-format/t031-000513_ca-01003.csv"
OK - it only has one file. Other datasets may have hundreds.
It’s is very simple to read the short-form data into a data frame
(well, a tibble actually). Make your request by copepid… be advised
some datasets have many many files and it can take a while to read then
all in.
x <- read_copepod(list_data(copepid = "ca-01003"), simplify = TRUE)
dplyr::glimpse(x)## Rows: 188
## Columns: 10
## $ shp_cruise <chr> "T031-000513", "T031-000513", "T031-000513", "T031-0005…
## $ date <dttm> 2000-05-13 00:00:04, 2000-05-13 00:00:04, 2000-05-13 0…
## $ lon <dbl> -63.53, -63.53, -63.47, -63.47, -63.41, -63.41, -63.31,…
## $ lat <dbl> 44.46, 44.46, 44.37, 44.37, 44.30, 44.30, 44.17, 44.17,…
## $ zupper <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
## $ zlower <dbl> 49, 49, 48, 48, 56, 56, 52, 52, 56, 56, 46, 46, 51, 51,…
## $ value_per_vol <dbl> 0.3640, 162.9663, 0.5809, 268.9008, 0.3050, 216.9040, 0…
## $ value_per_area <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
## $ lifestage <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
## $ lifestage_name <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
Alternatively, you can read the data in as a sf POINT object.
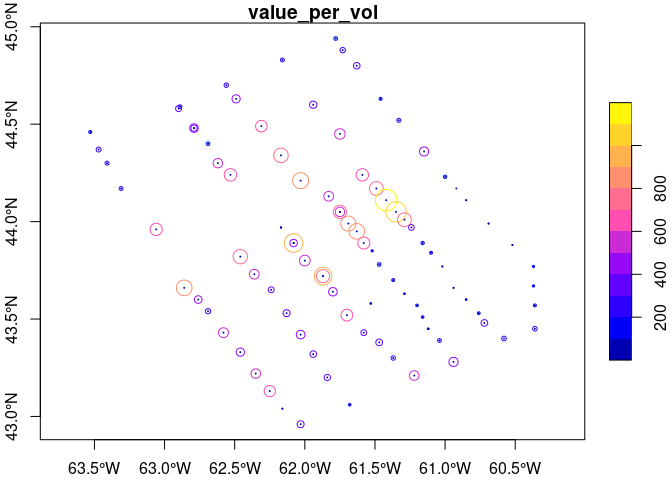
x <- read_copepod(list_data(copepid = "ca-01003"), simplify = TRUE, form = 'sf')
r <- range(x$value_per_vol)
cex <- (x$value_per_vol - r[1])/(r[2]- r[1]) * 3
plot(x['value_per_vol'], axes = TRUE, cex = cex)