Conveniently take IGV snapshots in multiple bam files over mutliple regions.
- The standard way of running
igver.pyis through docker or singularity. - Supports genomes listed in the #hosted-genome-list from the IGV team.
- IGVer isn't battle-hardened at all; any help/push/feedback will greatly help improving it! 🙏
igver.py --helpgives:
usage: igver.py [-h] --bam BAM [BAM ...] -r REGIONS -o OUTDIR [-g GENOME]
[-t TAG] [-mph MAX_PANEL_HEIGHT] [-od OVERLAP_DISPLAY]
[--overwrite] [-d IGV_DIR] [--config CONFIG]
Create temporary batchfile and run IGV for a region list
optional arguments:
-h, --help show this help message and exit
--bam BAM [BAM ...] Input tumor bam file(s) to be shown vertically
-r REGIONS, --regions REGIONS
Either a 'chr:start-end' string, or input regions file
with region columns to be shown horizontally
-o OUTDIR, --outdir OUTDIR
Output png directory
-g GENOME, --genome GENOME
Genome version [default: 'GRCh37']
-t TAG, --tag TAG Tag to suffix your png file [default: 'tumor']
-mph MAX_PANEL_HEIGHT, --max_panel_height MAX_PANEL_HEIGHT
Max panel height [default: 200]
-od OVERLAP_DISPLAY, --overlap_display OVERLAP_DISPLAY
'expand', 'collapse' or 'squish'; [default: 'squish']
--overwrite Overwrite existing png files [default: False]
-d IGV_DIR, --igv_dir IGV_DIR
/path/to/IGV_x.xx.x
--config CONFIG Additional preferences [default: None]- You can plug in additional IGV preferences as in https://github.com/igvteam/igv/wiki/Batch-commands -- an example would be
test/tag_haplotype.batch:
group TAG HP
colorBy TAG rl
sort READNAME
- AFAIK, the only way to modify the batch screenshot width is by modifying your
${IGV_DIR}/prefs.propertiesfile. There is a line that looks something likeIGV.Bounds=0,0,640,480, meaning that IGV set the bounds of the left, top, width, height (refer to igvteam/igv#161). I've tried to override this but seems that it doesn't work that way. For the example below, I've fixed my prefs.properties file so that the screenshot width is 800 (i.e. setIGV.Bounds=0,0,800,480).
- An example command getting two bam files as inputr, displayed vertically in the order put in (i.e. top panel:
haplotag_tumor.bam, bottom panel:haplotag_normal.bam), is as follows. - Here,
test/tag_haplotype.batchincludes additional IGV preferences to group and color haplotagged reads, as written above.
singularity run -B /juno docker://shahcompbio/igv igver.py \
--bam test/test_tumor.bam test/test_normal.bam \
-r test/region.txt \
-o test/snapshots \
-mph 500 -od squish \
--config test/tag_haplotype.batch- The regions file for the test case,
test/region.txt, includes 4 lines of different regions. The number of regions in the same line will lead to a snapshot with the regions horizontally aligned. You can annotate the region with an optional final field, which you can omit. - Here's the content of
test/region.txtand some explanation below.
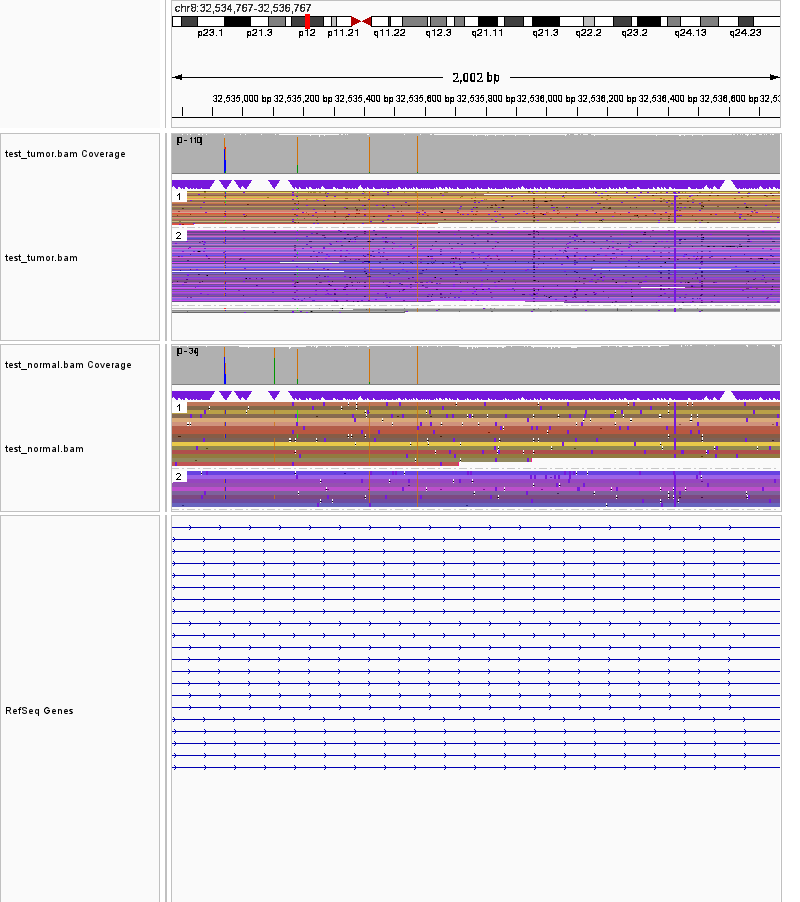
8:32534767-32536767 region_of_interest
8:32534767-32536767 19:11137898-11139898 translocation
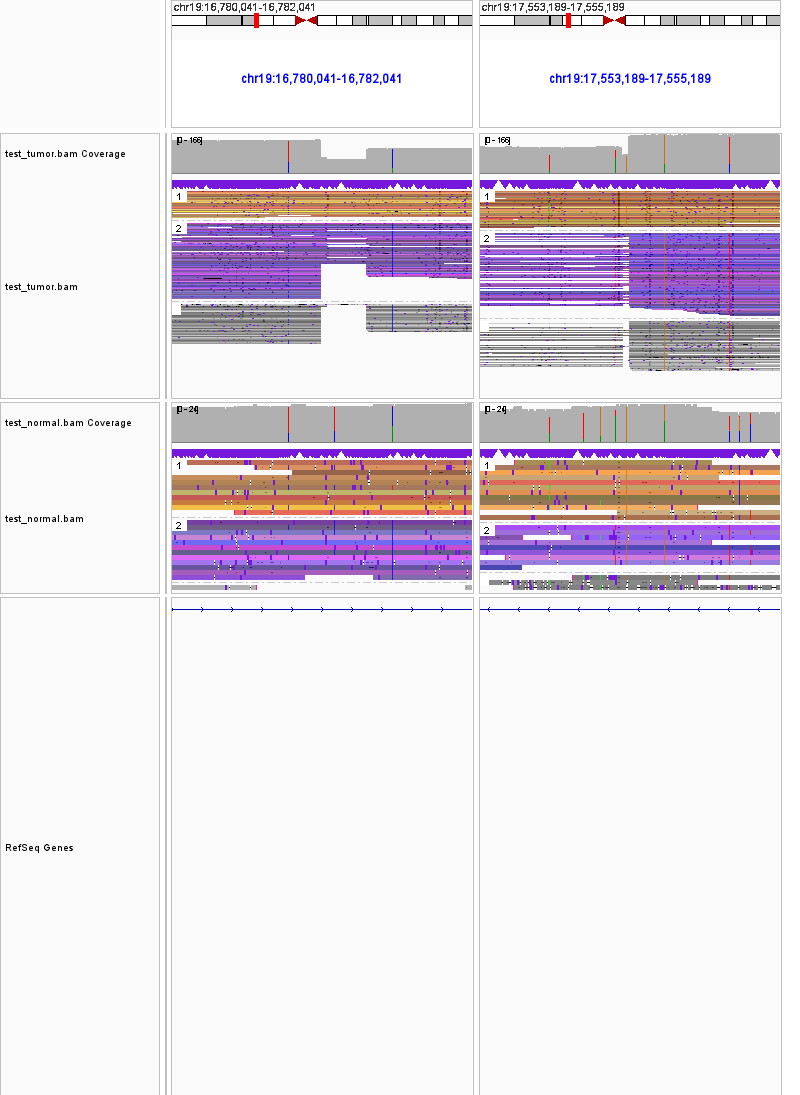
19:16780041-16782041 19:17553189-17555189 inversion
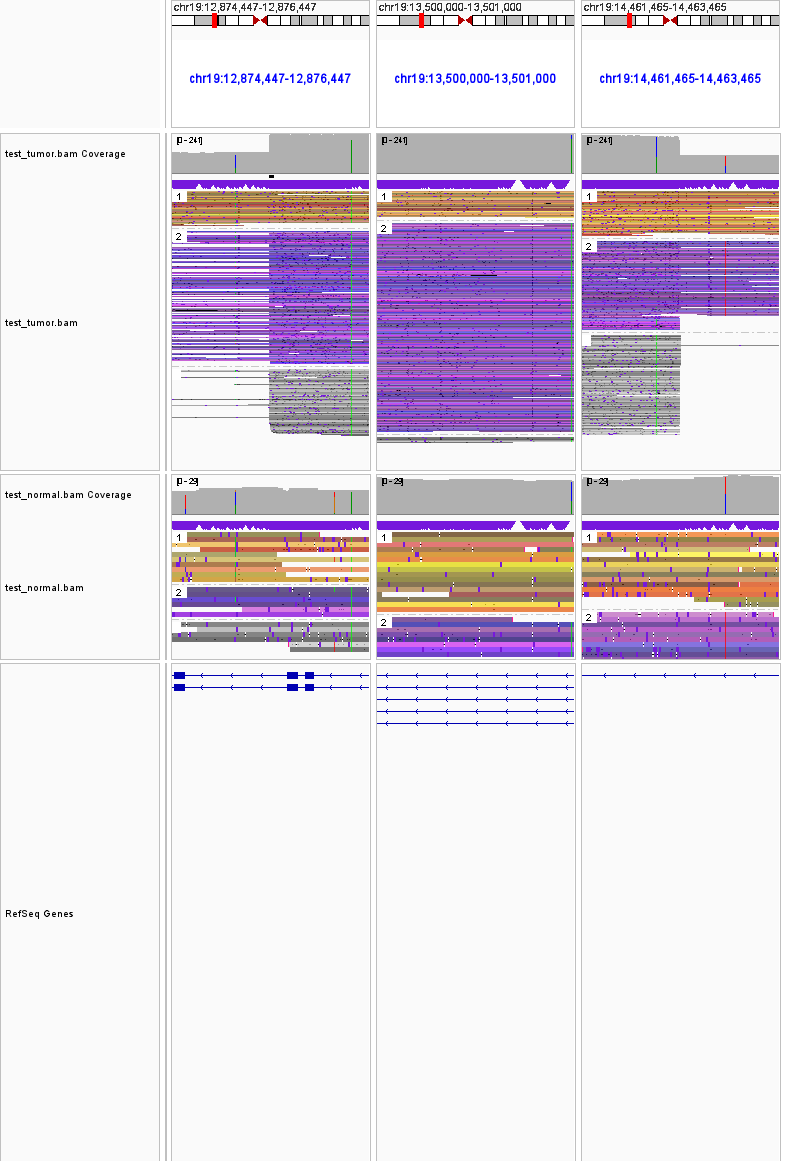
19:12874447-12876447 19:13500000-13501000 19:14461465-14463465 duplication
- The first region will take a 1001bp;1001bp snapshot on the region coined "region of interest", and create a png file
8-32534767-32536767.region_of_interest.tumor.pngin the OUTDIR. - The second region will take a 1001bp;1001bp snapshot on the two breakpoints of the translocation, and create a png file
8-32534767-32536767.19-11137898-11139898.translocation.tumor.pngin the OUTDIR. - The third region will take a 1001bp;1001bp snapshot on the two breakpoints of the inversion, and create a png file
19-16780041-16782041.19-17553189-17555189.inversion.tumor.pngin the OUTDIR. - The fourth region will take a 1001bp;1001bp;1001bp snapshot on the two breakpoints and a region inbetween, and create a png file
19-12874447-12876447.19-13500000-13501000.19-14461465-14463465.duplication.tumor.pngin the OUTDIR.
- You can see that the png files in OUTDIR includes
.tumoras a suffix. This is because the default TAG of the--tag / -toption is "tumor". You can set it to "None" to omit tagging the suffix.
- You can see the IGV snapshots already taken using the script above in
test/snapshots.