getting-started-bigquery
The repository contains examples of using BigQuery with genomics data. The code within each language-specific folder demonstrates the same set of queries upon the Platinum Genomes dataset. For more detail about this data see Google Genomics Public Data.
For more advanced examples, see BigQuery Examples
All languages will require a Project ID from a project that has the BigQuery API enabled.
- Follow the BigQuery sign up instructions if you do not yet have a valid project. (Note: you do not need to enable billing for the small examples in this repository)
- You can find the Project ID for your new project in the Google Developers Console.
Using the BigQuery browser tool
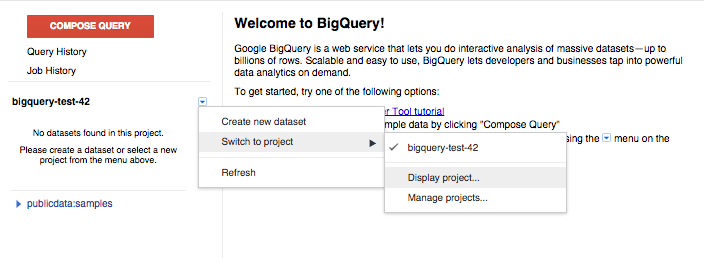
Instead of using code to call the BigQuery API, you can also use the Browser Tool to manually execute queries.
- Go to the BigQuery Browser Tool.
- Click on "Compose Query".
- Copy and paste the following query into the dialog box and click on "Run Query":
SELECT
reference_name,
COUNT(reference_name) AS num_records,
COUNT(call.call_set_name) AS num_calls
FROM
[genomics-public-data:platinum_genomes.variants]
GROUP BY
reference_name
ORDER BY
reference_name
View the results!
###Adding datasets from Google Genomics Public Data
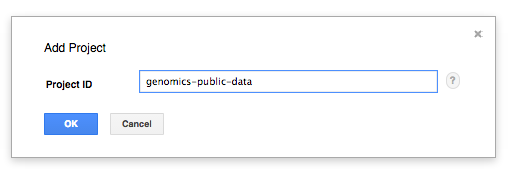
You can also add the Google Genomics Public Data BigQuery datasets to the browser tool so that they show up in the left-hand navigation pane.
- Click on the drop down icon beside your project name in the left-hand navigation pane.
- Pick ‘Switch to project’ in the menu, and ‘Display project...’ in the submenu
What next?
- New to BigQuery?
- See the query reference.
- See the BigQuery book Google BigQuery Analytics
- New to working with variants?
- See an overview of the VCF data format.
- Looking for more sample queries?
- See BigQuery Examples.