TMExplorer (Tumour Microenvironment Explorer) is a curated collection of scRNAseq datasets sequenced from tumours. It aims to provide a single point of entry for users looking to study the tumour microenvironment gene expressions at the single-cell level.
Users can quickly search available datasets using the metadata table, and then download the datasets they are interested in for analysis. Optionally, users can save the datasets for use in applications other than R.
This package will improve the ease of studying the tumour microenvironment with single-cell sequencing. Developers may use this package to obtain data for validation of new algorithms and researchers interested in the tumour microenvironment may use it to study specific cancers more closely.
- Install from Bioconductor
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("TMExplorer")
- Install from GitHub for latest development release
library(devtools)
install_github("shooshtarilab/TMExplorer")
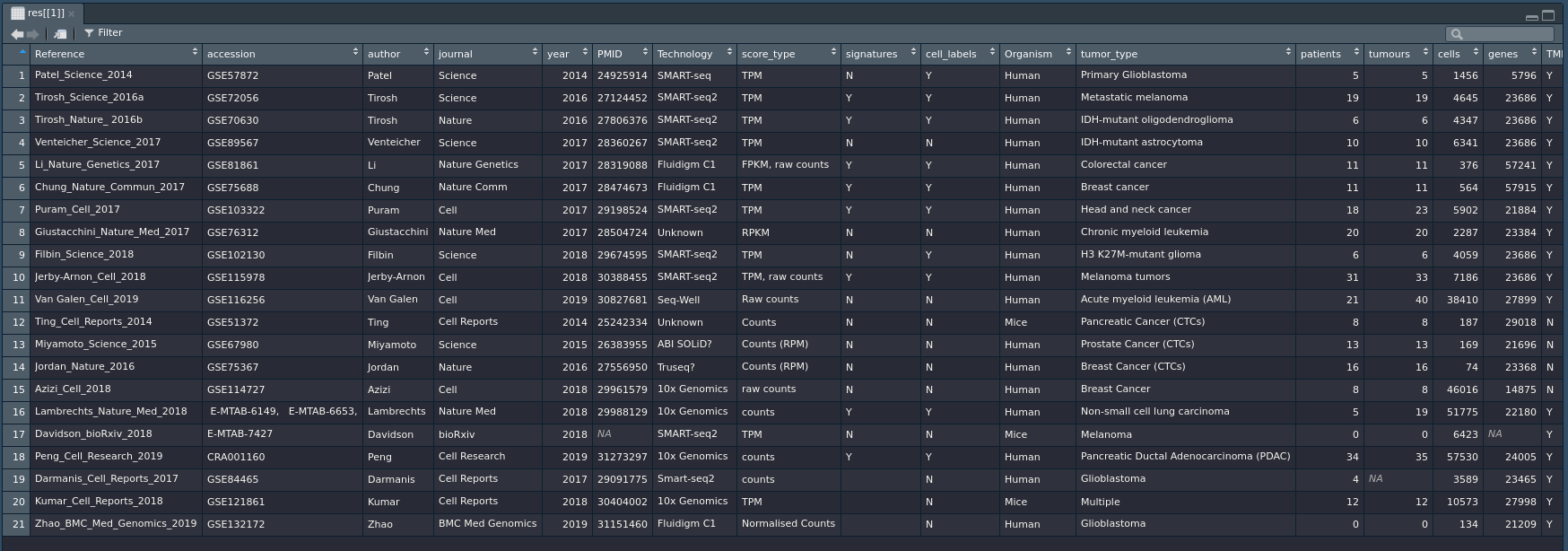
Start by exploring the available datasets through metadata.
> res = queryTME(metadata_only = TRUE)
This will return a list containing a single dataframe of metadata for all available datasets. View the metadata with View(res[[1]]) and then check ?queryTME for a description of searchable fields.
Note: in order to keep the function's interface consistent, queryTME always returns a list of objects, even if there is only one object. You may prefer running res = queryTME(metadata_only = TRUE)[[1]] in order to save the dataframe directly.
The metatadata_only argument can be applied alongside any other argument in order to examine only datasets that have certain qualities. You can, for instance, view only breast cancer datasets by using
> res = queryTME(tumour_type = 'Breast cancer', metadata_only = TRUE)[[1]]
| Search Parameter | Description | Examples |
|---|---|---|
| geo_accession | Search by GEO accession number | GSE72056, GSE57872 |
| score_type | Search by type of score shown in $expression | TPM, RPKM, FPKM |
| has_signatures | Filter by presence of cell-type gene signatures | TRUE, FALSE |
| has_truth | Filter by presence of cell-type labels | TRUE, FALSE |
| tumour_type | Search by tumour type | Breast cancer, Melanoma |
| author | Search by first author | Patel, Tirosh, Chung |
| journal | Search by publication journal | Science, Nature, Cell |
| year | Search by year of publication | <2015, >2015, 2013-2015 |
| pmid | Search by PubMed ID | 24925914, 27124452 |
| sequence_tech | Search by sequencing technology | SMART-seq, Fluidigm C1 |
| organism | Search by source organism | Human, Mice |
| sparse | Return expression in sparse matrices | TRUE, FALSE |
In order to search by single years and a range of years, the package looks for specific patterns. '2013-2015' will search for datasets published between 2013 and 2015, inclusive. '<2015' will search for datasets published before or in 2015. '>2015' will search for datasets published in or after 2015.
Once you've found a field to search on, you can get your data.
> res = queryTME(geo_accession = "GSE72056")
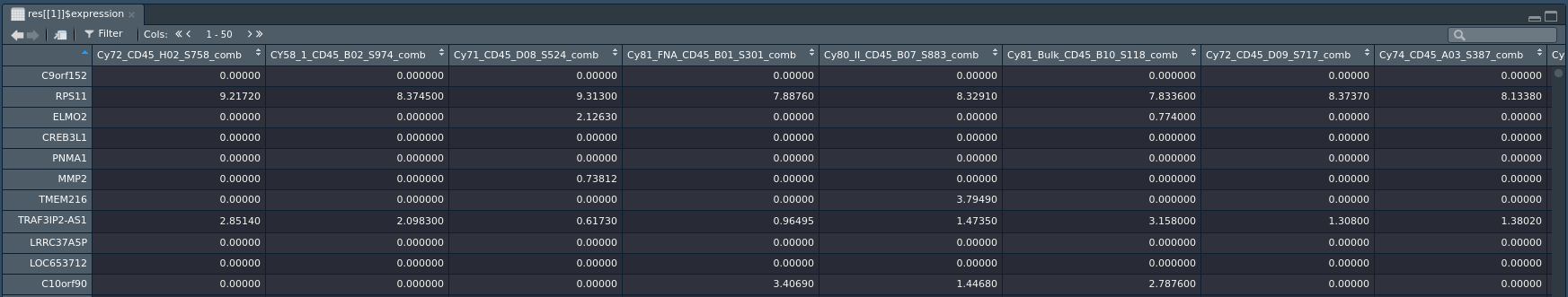
This will return a list containing dataset GSE72056. The dataset is stored as a SingleCellExperiment object, with the following metadata list:
| Attribute | Description |
|---|---|
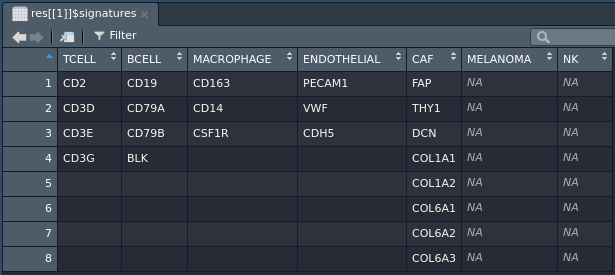
| signatures | A data.frame containing the cell types and a list of genes that represent that cell type |
| cells | A list of cells included in the study |
| genes | A list of genes included in the study |
| pmid | The PubMed ID of the study |
| technology | The sequencing technology used |
| score_type | The type of score shown in tme_data$expression |
| organism | The type of organism from which cells were sequenced |
| author | The first author of the paper presenting the data |
| tumour_type | The type of tumour sequenced |
| patients | The number of patients included in the study |
| tumours | The number of tumours sampled by the study |
| geo_accession | The GEO accession ID for the dataset |
To access the expression data for a result, use
> View(counts(res[[1]]))
Cell type labels are stored under colData(res[[1]]) for datasets for which cell type labels are available.
To access metadata for a dataset, use
> metadata(res[[1]])
Specific metadata entries can be accessed by specifying the attribute name, for instance
> metadata(res[[1]])$pmid
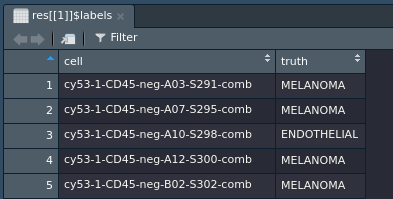
Say you want to measure the performance of cell-type classification methods. To do this, you need datasets that have the true cell-types available.
> res = queryTME(has_truth = TRUE)
This will return a list of all datasets that have true cell-types available. You can see the cell types for the first dataset using the following command:
> View(colData(res[[1]]))
The first column of this dataframe contains the cell barcode, and the second contains the cell type.
Some cell-type classification methods require a list of gene signatures, to return only datasets that have cell-type gene signatures available, use:
> res = queryTME(has_truth = TRUE, has_signatures = TRUE)
> View(metadata(res[[1]])$signatures)
To facilitate the use of any or all datasets outside of R, you can use saveTME(). saveTME takes two parameters, one a tme_data object to be saved, and the other the directory you would like data to be saved in. Note that the output directory should not already exist.
To save the data from the earlier example to disk, use the following commands.
> res = queryTME(geo_accession = "GSE72056")[[1]]
> saveTME(res, '~/Downloads/GSE72056')
[1] "Done! Check ~/Downloads/GSE72056 for files"
The result is three CSV files (gene expressions, cell labels, and gene signatures) that can be used in other programs. In the future we will support saving in other formats.
NOTE: saveTME is currently not compatible with sparse datasets. This is due to the size of some datasets and the memory required to convert them to a dense matrix that can be written to a csv file. To save the elements of a sparse object, use write.table() and as.matrix(counts(res)), keeping in mind that doing this with some of the larger datasets may cause R to crash.
While many of the datasets included in this package are small enough to be loaded and stored, even as dense matrices, on machines with an 'average' amount of memory (8-32gb), there are a few larger datasets that cannot be fully manipulated without a significant amount of memory. With this in mind, we recommend using sparse = TRUE when possible and using a system with at least 64gb of RAM for full functionality.
If you are experience crashes due to memory limitations, try using sparse = TRUE or grabbing datasets individually using the geo_accession parameter.
The following is a list of datasets that can not be converted between sparse and dense formats on a personal machine (Ryzen 5 3600, 16gb RAM)
- Van Galen, Cell 2019, GSE116256
- Azizi, Cell 2018, GSE114727
- Lambrechts, Nature Med 2018, E-MTAB-6149
- Davidson, bioRxiv 2018, E-MTAB-7427
- Peng, Cell Research 2019, CRA001160