Bassel Ghaddar 7/31/2022
SAHMI enables the systematic recovery and denoising of microbial signals from genomic sequencing of host tissues. The pipeline consists of R command line functions that implement the following main steps which are further described below.
- Taxonomic classification (recommended with Kraken2Uniq)
- Extract microbiome reads
- Single-cell k-mer analysis
- Barcode level signal denoising (barcode k-mer correlation test)
- Sample-level signal denoising (sample k-mer correlation tests)
- Identifying contaminants and false positives (cell line quantile test)
- Quantitation of microbes and creating the barcode-metagenome counts matrix
- Joint analysis of host and microbial data
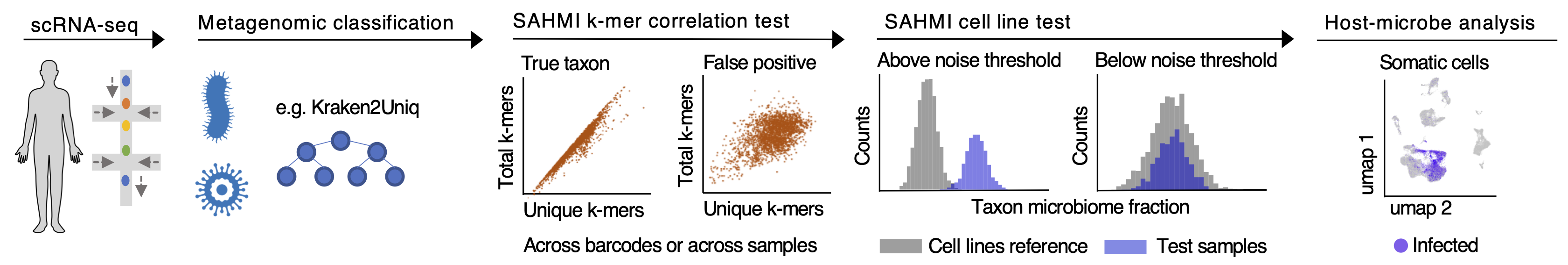 Figure 1A. A schematic representation of the SAHMI workflow
Figure 1A. A schematic representation of the SAHMI workflow
Please see the references below for more information.
Please contact Bassel Ghaddar (bassel.ghaddar@gmail.com) or Subhajoyti De (subhajyoti.de@rutgers.edu) for any questions.
No installation of SAHMI is required - all scripts run from the command line as described. Note that for all file paths that are input as function arguments the path must end in a backslash. Scripts can be made executable by running
chmod +x script.R
Rscript script.R -hThe following R packages must be installed prior to running some SAHMI functions: optparse, stringr, tidyverse, dplyr, ShortRead, data.table
Metagenomic classification of paired-end reads from single-cell RNA sequencing fastq files can be performed using any k-mer based mapper that identifies a taxonomic ID for each k-mer and read. However, SAHMI is optimized to run with Kraken2Uniq, which finds exact matches of candidate 35-mer genomic substrings to the lowest common ancestor of genomes in a reference metagenomic database. It is essential that all realistically possible genomes are included as mapping references at this stage (e.g. host, known vectors, etc.), or that host mappable reads are excluded. The required outputs from this step are: a Kraken summary report with sample level metagenomic counts, a Kraken output file with read and k-mer level taxonomic classifications, an MPA-style report, and raw sequencing fastq files with taxonomic classification for each read. Please see https://github.com/DerrickWood/kraken2/blob/master/docs/MANUAL.markdown for more details on installation and usage of Kraken2/KrakenUniq.
The script run_kraken.r is included for convenience of running Kraken2Uniq and creating the MPA style report and other systematically named Kraken outputs.
run_kraken.r
--samplesample name--fq1path to fastq 1 file--fq2path to fastq 2 file--out_pathoutput directory path--ncbi_blast_pathpath to ncbi-blast (see Kraken documentation for details)--Kraken2Uniq_pathpath to Kraken2 main 'kraken2' function--kraken_database_pathpath to kraken database--kreport2mpa_pathpath to kreport2mpa.py function (included in SAHMI/functions)--pairedare fastq files paired end (T) or single-end/unpaired (F). Default is T.
The output includes fastq files with Kraken NCBI taxonomic assignments for each read, an output file containing k-mer level taxonomic data, and Kraken standard, uniq, and MPA style reports.
Microbiome reads next need to be extracted from the fastq files and the Kraken output file using the following scripts:
extract_microbiome_reads.r
--sample_namesample name--fqpath to classified fastq file (sample_1.fq/sample_2.fq) (if paired end reads this function must be once for each read)kraken_reportpath to kraken2uniq report (sample.kraken.report.txt)mpa_reportpath to mpa style report (sample.kraken.report.mpa.txt)out_pathoutput path
extract_microbiome_output.r
--sample_namesample name--output_filepath to kraken output file (sample.output.txtkraken_reportpath to kraken2uniq report (sample.kraken.report.txt)mpa_reportpath to mpa style report (sample.kraken.report.mpa.txt)out_pathoutput path
The outputs are fasta files for microbiome reads and a microbiome output file.
The next step is tabulating k-mer statistics across individual barcodes using the script sckmer.r for paired end data and sckmer_unpaired.r for unpaired/single-end sequence data. These functions count the number of k-mers and unique k-mers assigned to a taxon across barcodes. The cell barcode and unique molecular identifier (UMI) are used to identify unique barcodes and reads. Data is reported for taxa of pre-specified ranks (default genus + species) taking into account all subsequently higher resolution ranks. Reads with any k-mers mapped to the host (e.g. human) are discarded. Reads with >50% of the k-mers map outside the taxon's lineage are also discarded. The output is a table of barcodes, taxonomic IDs, number of k-mers, and number of unique k-mers.
sckmer.r
--sample_namesample name--fa1path to microbiome fasta 1 file (sample_1.fa)--fa2path to microbiome fasta 2 file (sample_2.fa)--microbiome_output_filepath to microbiome output file (sample.microbiome.output.txt)kraken_reportpath to kraken2uniq report (sample.kraken.report.txt)mpa_reportpath to mpa style report (sample.kraken.report.mpa.txt)out_pathoutput pathcb_lennucleotide length of cell barcodesumi_lennucleotide length of unique molecular identifiers (UMI)rankstaxa ranks to analyze. more than one value allowed (e.g. c('G', 'S')). See Kraken report for more detailhosthost taxonomy ID to excludemin_fracminimum fraction of k-mers directly assigned to taxon ID or its lineage to use read.nsamplemax number of barcodes to sample per taxon ID
Note that parameters for sckmer_unpared.r are the same but do not include fa2.
True taxa are detected on multiple barcodes and with a proprotional number of total and unique k-mer sequences across barcodes, measured as a significant Spearman correlation between the number of total and unique k-mers across barcodes. We demonstrate this using example data from Zhang et al., Cell Reports 2019 for a gastric metaplasia sample positive for Helicobacter pylori. Running SAHMI steps 1-3 generates the Kraken report and sckmer.txt.
library(dplyr)
library(tidyverse)
# kraken report
report = read.delim('./example data/SRR9713132.kraken.report.txt', header = F)
report$V8 = trimws(report$V8)
report[report$V8 %in% c('Homo sapiens', 'Bacteria', 'Fungi', 'Viruses'), ]
# sckmer data
kmer_data = read.table('./example data/SRR9713132.sckmer.txt', header = T)
head(kmer_data)We see that Kraken intially detected a large number of unique genera and species in the sample:
length(unique(report$V8[report$V6 %in% c('G', 'S')])) However, only a small subset of unique taxonomy IDs were detected on sufficient barcodes to test the Spearman corrletion:
# barcode k-mer correlation tests on taxonomy IDs detected on >3 barcodes and with >1 k-mer
# taxid = NCBI taxonomic ID, r = Spearman correlation coefficient, p = adjusted p-value
c = kmer_data %>%
subset(kmer > 1) %>%
group_by(taxid) %>%
mutate(nn = n()) %>%
subset(nn > 3) %>%
group_by(taxid) %>%
summarize(r = cor.test(kmer, uniq, method = 'spearman')$estimate,
p = cor.test(kmer, uniq, method = 'spearman')$p.value,
.groups='keep') %>%
mutate(p = p.adjust(p))
c$name = report$V8[match(c$taxid, report$V7)] # add taxa names
cThis test significantly filters list of possible taxa and enriches for real taxa in an individual sample.
In the low-microbiome biomass setting, real microbes also exhibit a proportional number of total k-mers, number of unique k-mers, as well as number of total assigned sequencing reads across samples; i.e. the following three Spearman correlations are significant when tested using sample-level data provided in Kraken reports: cor(#reads, #k-mers), cor(#reads, #unique k-mers), and cor(#k-mers, #unique k-mers). We demonstrate this using data from 32 samples from Zhang et al. 2019 (8 H.pylori+, 24 H. pylori-).
The R function read_kraken_reports.r is provided for conveinence of reading and formatting multiple Kraken reports. Note that it is not a command line function and needs to be executed in R prior to using it.
read_kraken_reports.r Arguements:
- files, vector of standard Kraken report file paths (sample.kraken.txt)
- sample_names, optional sample name
- study_name, optional study name
- min_reads, filter taxa with > (default 2) assigned reads
- min_uniq, filter taxa with > (default 2) unique k-mers
Kraken reports for this study are included as an RDS object.
kr = readRDS('./example data/zhang.reports.RDS')
krColumn names key: taxid, NCBI taxonomy ID, reads, # reads assigned; min, estimated # minimizers (k-mers), see Kraken documentation for more detail; uniq, estimated # unique minimizers (k-mers); rpm, reads per million; rpmm, reads per million microbiome reads.
Running the three sample-level k-mer correlation tests on genus and species resolution taxa yields:
require(ggplot2)
# remove taxa detected in < 3 samples
kr = kr %>%
group_by(taxid) %>%
mutate(nn = n()) %>%
subset(nn > 2) %>%
select(-nn)
# run correlations
c2 = kr %>%
subset(rank %in% c('G', 'S')) %>%
group_by(name) %>%
summarize(r1 = cor(min,uniq,method='spearman'),
r2 = cor(min,reads,method='spearman'),
r3 = cor(reads,uniq,method='spearman'),
p1 = cor.test(min,uniq,method='spearman')$p.value,
p2 = cor.test(min,reads,method='spearman')$p.value,
p3 = cor.test(reads,uniq,method='spearman')$p.value
)
c2
# making a scatter plot of correlation test results.
# Each point is a taxon. x-axis, Spearman correlation value between #k-mers vs. #unique k-mers; y-axis, correlation value between #k-mers vs. #reads; color, correlation value between #reads vs. #unique k-mers. Lines represent contour density.
ggplot(c2, aes(r1, r2, color = r3))+
geom_point() +
geom_density_2d(size = 0.75, colour = "black") These results show a wide range of values for the three correlations, with only a subset of taxa having signifcant values in all three metrics.
The previous steps enrich for true taxa that produced diverse RNA. The next step in SAHMI is to identify contaminant taxa and spurious false positive assignments. These can be identified based on the widely observed pattern that contaminants appear at higher frequencies in low concentration or negative control samples. In the absence of experimentally matched negative controls, SAHMI provides a negative control resource comprised of microbiome profiles from 2,491 sterile cell experiments from around the world. For each taxon in a test sample, SAHMI compares the fraction of microbiome reads assigned to the taxon [i.e. taxon counts/sum(all bacterial, fungal, viral counts), in reads per million] to the microbiome fraction assigned to the taxon in all cell line experiments. Using the microbiome fraction comparison normalizes for experiments having a varying number of total sequencing reads or varying underlying contamination. SAHMI tests whether the taxon microbial fraction in the test sample is > 99th percentile (by default) of the taxon’s microbiome fraction distribution in cell line data using a one-sample quantile test. Taxa whose counts fall within the cell line distribution are identified as below the cell-line noise threshold. Users may choose how stringently to select the quantile threshold for significance testing.
Table S4.xlsx contains the reads per million microbiome reads (rpmm) percentile data for taxa detected in cell lines. Each row corresponds to a taxon which is identified by name, rank (rank, G = genus, S = species), and NCBI taxonony id (taxid). The columns with numeric names (0.01-1) correspond to rpmm percentile values in the cell lines dataset. For example, the column "0.01" contains the rpmm first percentile value for each taxon, "0.5" contains the 50th percentile data, etc. These rpmm percentile values can be directly compared to the rpmm values in the test samples. Users may also wish to work with the raw cell lines genus and species resolution data contained in cell.lines.txt (Dropbox link below) to get a better feel for taxa distributions or to include or exclude zero counts in quantile calculations. Percentile values in Table S4.xlsx are calculated only from samples in which a taxon was reported with >2 reads and >2 unique k-mers. Users may wish to include all reported samples or zero-count samples.
To denoise and decontaminate the gastric metaplasia samples from above (SRR9713132) we combined data from the barcode and sample k-mer correlation tests and keep taxa that pass all tests. We then compare the remaining taxa counts to their distribution in the cell line data. The same sample-level data would be used for other samples in the study, but each sample would have its own sckmer results.
# combine k-mer correlation tests, filter for significant values and species resolution
c3 = left_join(c, c2, by = 'name') %>%
left_join(select(kr, rank, name) %>% distinct()) %>%
subset(r1>0 & r2>0 & r3>0 & p<0.05 & p1<0.05 & p2<0.05 & p3<0.05 & rank == 'S')
c3For the taxa identified, we combined their data (for all samples) with the cell line data and plot density plots of their reads per million microbiome reads.
The raw cell lines microbiome data can be downloaded here:
https://www.dropbox.com/s/r6xvw1589lqyqts/cell.lines.txt?dl=0
library(scales)
# cell.lines = readxl::read_xlsx('Table S4.xlsx')
cell.lines = read.delim('/Users/bassel/Downloads/cell.lines.txt', header = T) %>% tibble()
df = cell.lines[,1:11] %>% mutate(study = 'cell lines'); df = df[, -2]
df = rbind(df, kr)
ggplot(subset(df, name %in% c3$name), aes(rpmm, fill = study, ..scaled..)) +
geom_density(alpha = 0.5, color = NA) +
facet_grid(~name, scales='free') +
scale_fill_manual(values = c(`cell lines` = 'grey50', zhang = 'navyblue')) +
theme_minimal() +
xlab('Microbiome reads per million') +
ylab('Density') +
scale_x_log10(labels = trans_format("log10", math_format(10^.x)),
breaks = trans_breaks("log10", function(x) 10^x, n=4),
oob = scales::squish, expand = c(0,0)) +
scale_y_continuous(expand = c(0,0)) +
theme(legend.title = element_blank(),
panel.border = element_rect(fill = NA, color = 'black'),
strip.background = element_blank(),
axis.ticks.x = element_line(size=0),
axis.ticks.y = element_blank(),
panel.grid = element_blank(),
strip.text = element_text(color = 'black', size = 10),
axis.text.x = element_text(color = 'black', size = 10),
axis.text.y = element_text(color = 'black', size = 10),
axis.title.y = element_text(color = 'black', size = 10),
axis.title.x = element_text(color = 'black', size = 10),
plot.margin = unit(c(0, 0.1, 0, 0), "cm"),
legend.key.size = unit(0.2, "cm"),
legend.text = element_text(color = 'black', size = 10),
legend.position = 'bottom')These density plots show that the only taxon detected at counts per million greater than found in the cell line data is Helicobacter pylori, and only in some samples (middle and right peaks). Quantitative results are obtaind by direclty comparing test sample rpmm to taxa quantile data from Table S4 or cell.lines.txt. Here we merge results from kraken report, k-mer correlation tests, and cell line quantile test for sample SRR9713132.
# get quantiles from cell line data (or alternatively use Table S4)
qtile = 0.99
q_df = cell.lines %>%
group_by(name, rank) %>%
summarize(CLrpmm = 10^quantile(log10(rpmm), qtile, na.rm = T),
.groups = 'keep')
left_join(c3, q_df, by = c('name', 'rank')) %>%
left_join(subset(kr, sample == 'SRR9713132') %>% select(name, rpmm), by = c('name'))After identifying true taxa, reads assigned to those taxa are extracted and then undergo a series of filters. The cell barcode and UMI are used to demultiplex the reads and create a barcode x taxa counts matrix. The full taxonomic classification of all resulting barcodes and the number of counts assigned to each clade are tabulated. The follwing command line R function performs this task:
taxa_counts.r
--sample_namesample name--fa1path to microbiome fasta 1 file (sample_1.fa)--fa2path to microbiome fasta 2 file (sample_2.fa)taxatsv file containing NCBI taxonomic IDs to extract (will summarize appropriate children data)kraken_reportpath to kraken2uniq report (sample.kraken.report.txt)mpa_reportpath to mpa style report (sample.kraken.report.mpa.txt)out_pathoutput pathcb_lennucleotide length of cell barcodesumi_lennucleotide length of unique molecular identifiers (UMI)
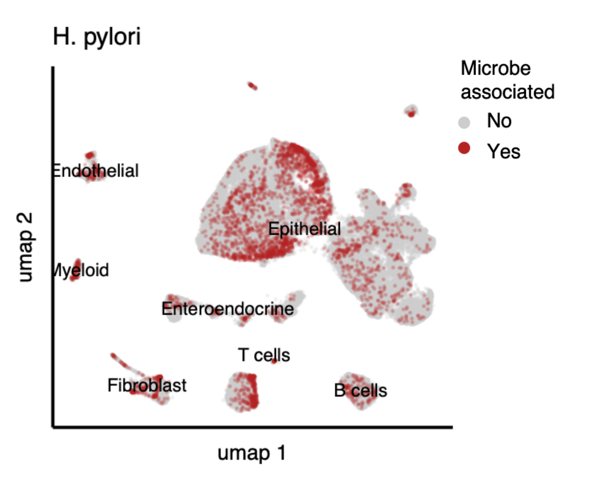
SAHMI produces a final barcode by metagenomic counts matrix which can be jointly analyzed with the somatic single-cell data. For starters, we can identify barcodes that tag both host and microbial RNA and visualize them on a UMAP plot.
Please see the following publications for more details:
Coming soon.