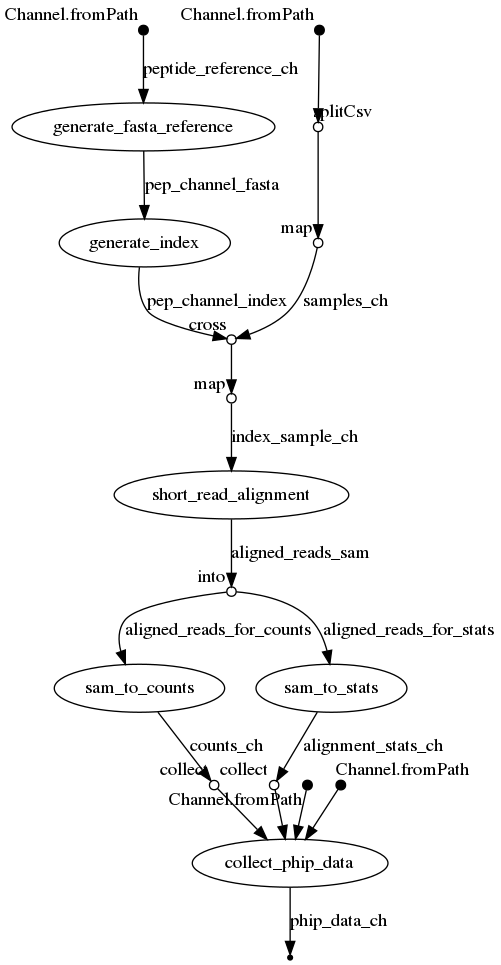
A general PhIP-Seq short read alignment pipeline using NextFlow to produce a dataset containing the raw alignment counts , sample annotation table, and peptide annotation table , all merged into an xarray DataSet. This dataset organization can subsequently be queried analyzed by using the phippery Python package.
This repository exists primarily to host the bleeding edge pipeline script, and should not need to be cloned unless one would like to modify the pipeline. If looking to simply run with the appropriate configuration scripts, one can simply use the Nextflow's build in git aware infrastructure
$ nextflow run matsengrp/phip-flow/PhIP-Flow.nf -C foo-phip.config -o bar.phip
For example configuration files and more helpful materials, the Documentation will walk you through examples that can be found in the Template repository
The pipeline Directed Acyclic Graph is visualized below: