SeuratV3Wizard: R Shiny interface for Seurat single-cell analysis library
New Features:
Sep 5, 2019
- all plots can be downloaded
- file with multiple samples can be uploaded
- tsne and umap are both always calculated
Apr 25, 2019
Online/Live instance:
You can try it online at http://nasqar.abudhabi.nyu.edu/SeuratV3Wizard
Pre-print:
NASQAR: A web-based platform for High-throughput sequencing data analysis and visualization
Run using docker (Recommended):
Make sure Docker (version >= 17.03.0-ce) is installed.
docker run -p 80:80 aymanm/seuratv3wizard
This will run on port 80
To run on a different port:
docker run -p 8083:80 aymanm/seuratv3wizard
This will run on port 8083
Local Install:
Make sure to have devtools installed first
devtools::install_github("nasqar/seuratv3wizard")
Optional: For ucsc cellbrowser support, make sure to follow the installation instructions here. For linux-based OS, type the following in the terminal:
sudo pip install cellbrowser=0.5.6
Run:
library(SeuratV3Wizard)
SeuratV3Wizard()
This will run on http://0.0.0.0:1234/ by default
To run on specific ip/port:
ip = '127.0.0.1'
portNumber = 5555
SeuratV3Wizard(ip,portNumber)
This will run on http://127.0.0.1:5555/
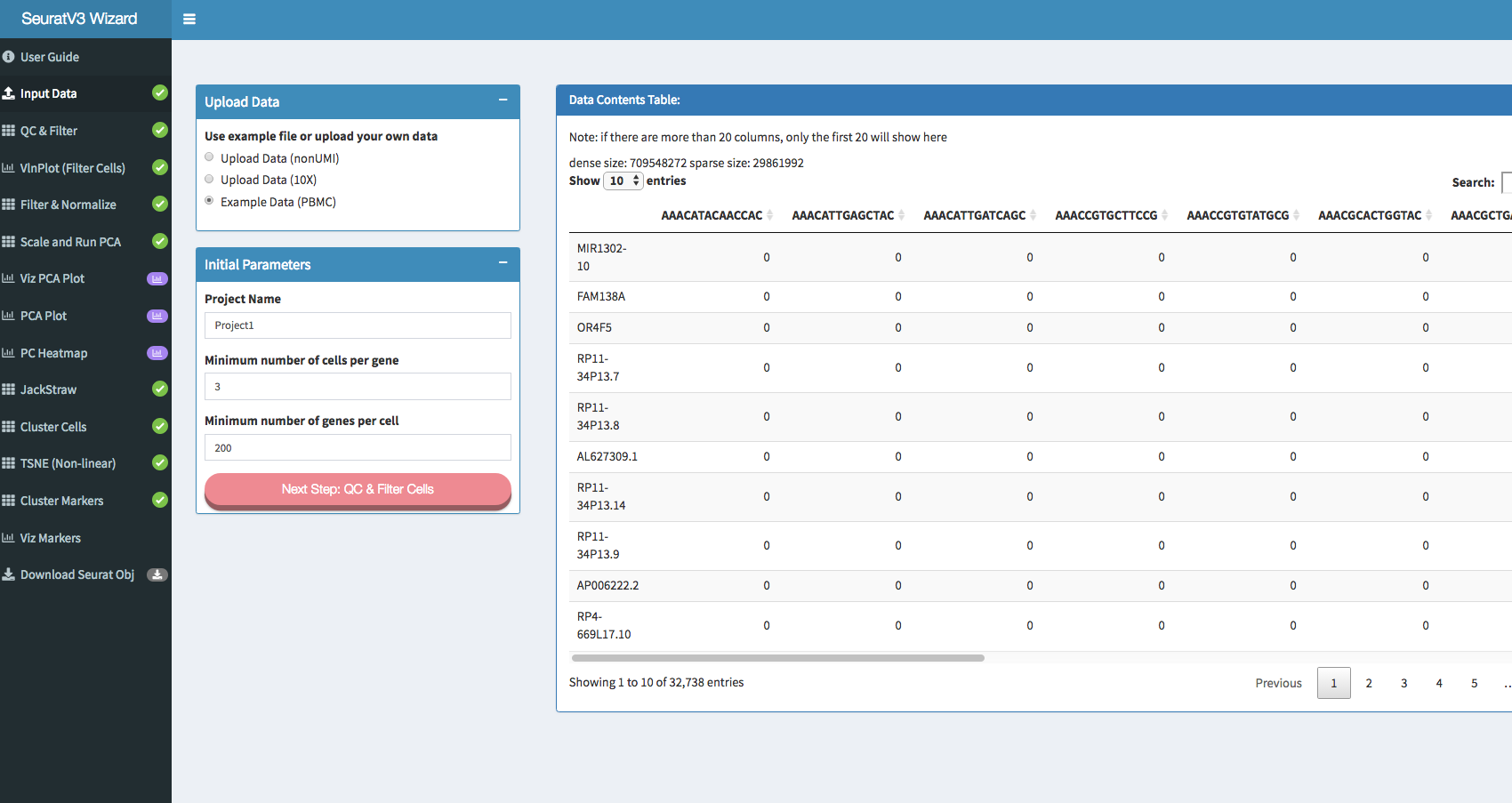
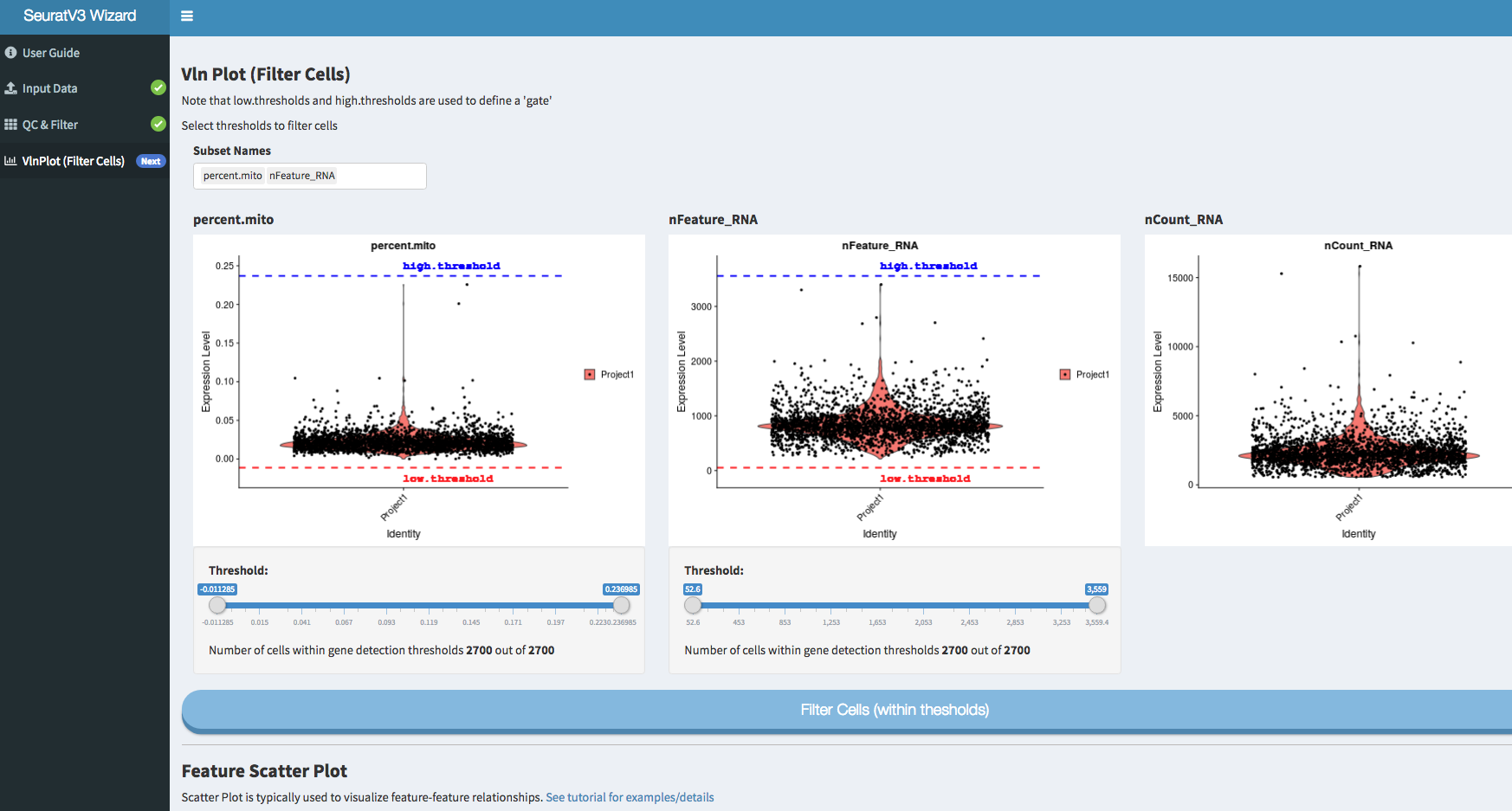
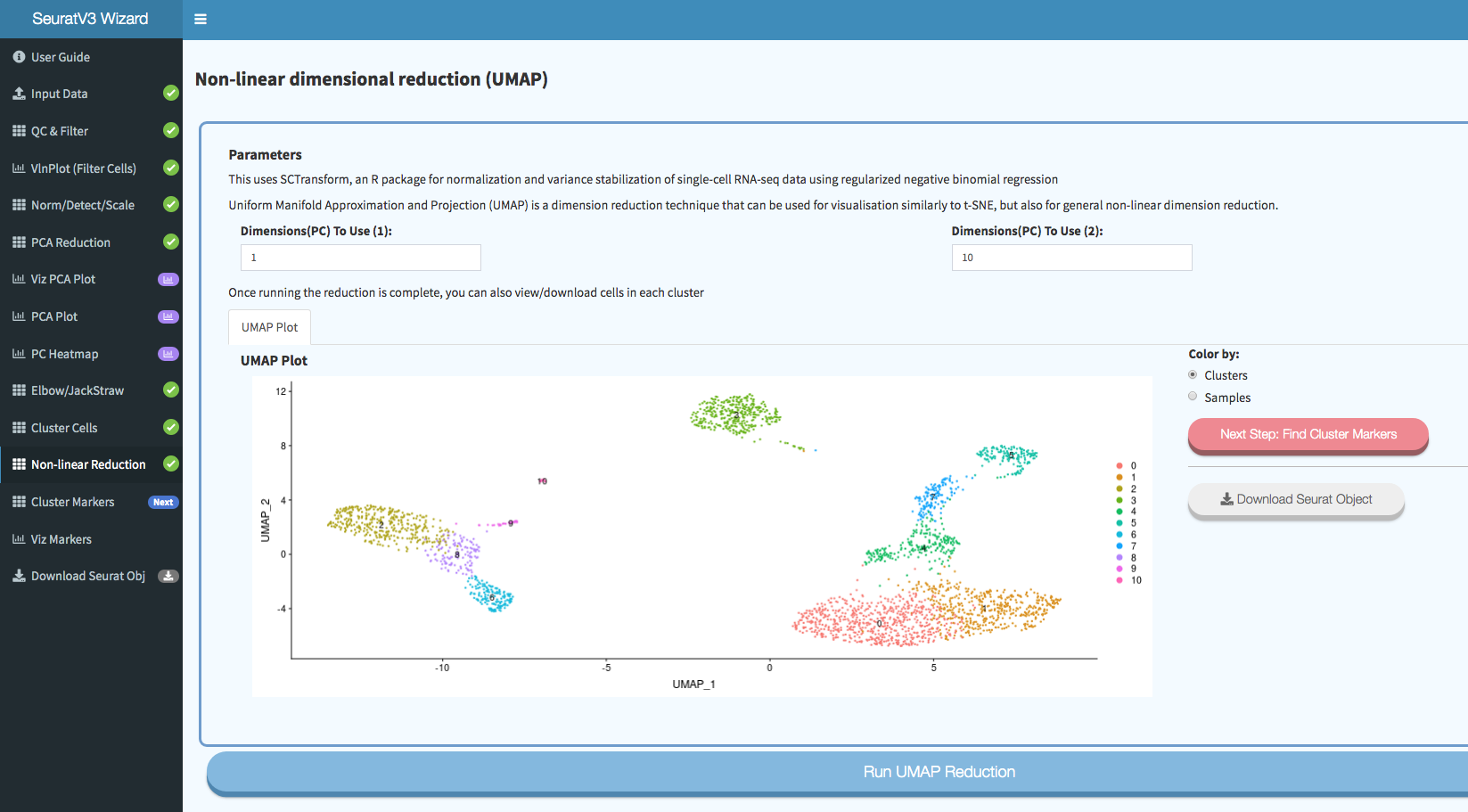
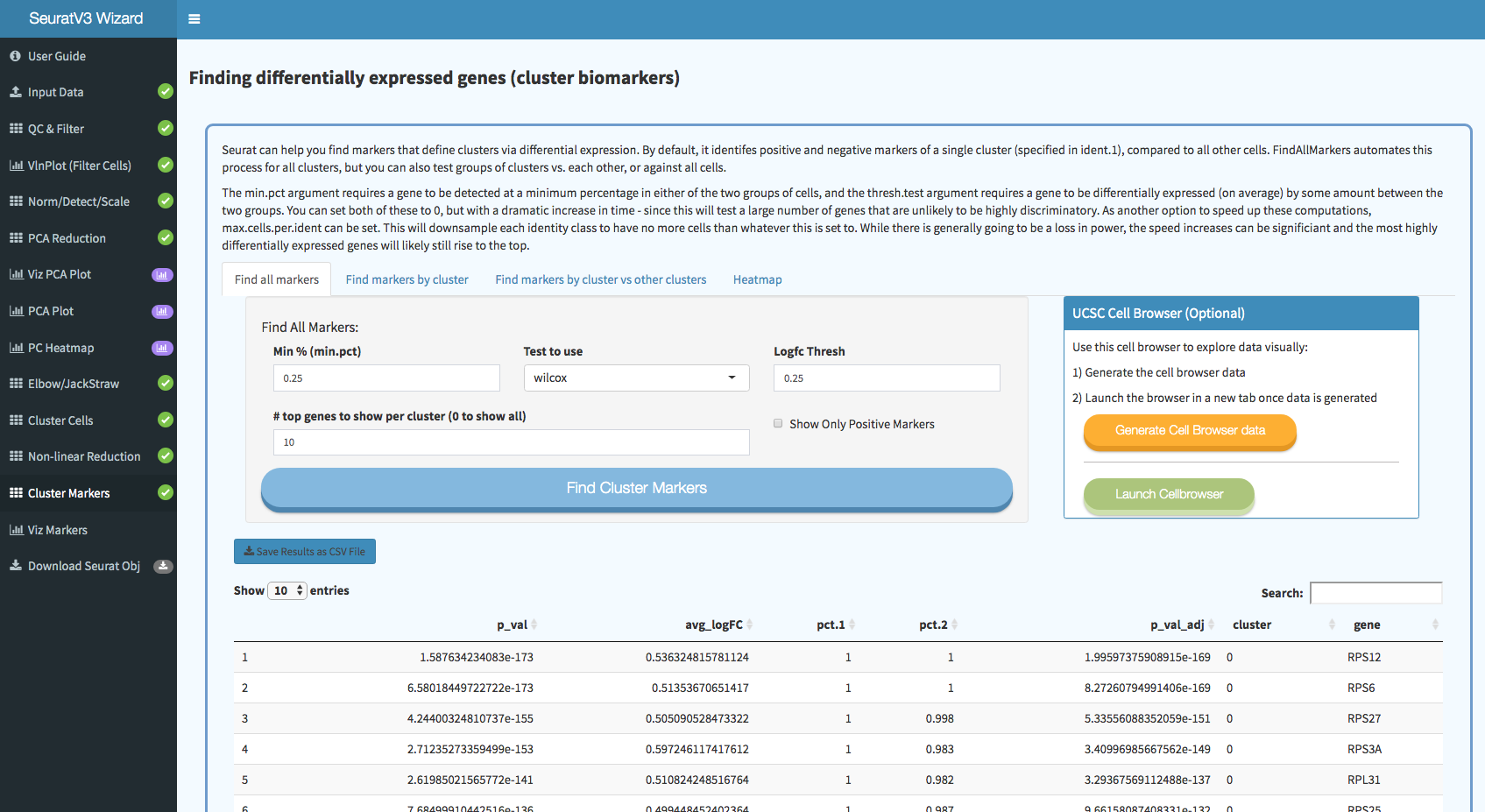
Screenshots:
Acknowledgements:
-
Rahul Satija, Andrew Butler and Paul Hoffman (2017). Seurat: Tools for Single Cell Genomics. R package version 2.2.1. https://CRAN.R-project.org/package=Seurat