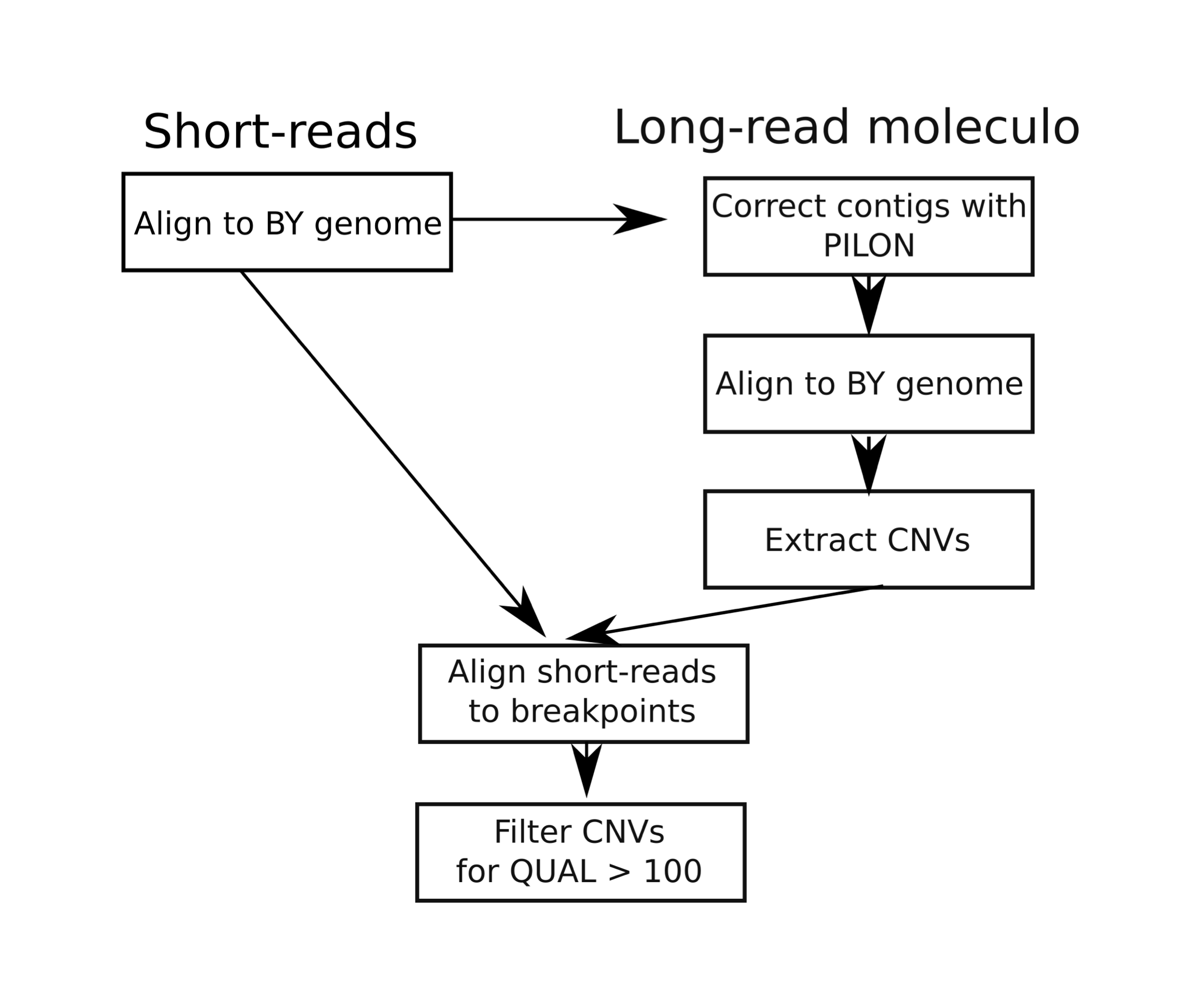
LRHCNV utilises long-reads and short-reads to call structural variants.
The long reads are used to uncover a set of putative CNVs, and then the short reads are used to provide support for the extracted breakpoints.
Simplied version of the pipeline utilised a moleculo genome assembly is shown below.
SAMPLE_NAME SCAFFOLDS PE SE
SM1 SM.fasta P1*.gz:P2*.gz,P1*.gz:P2*.gz fastq.gz