CBS-miRSeq
Availability and requirements
Project name: CBS-miRSeq: a robust comprehensive bioinformatics pipeline for microRNA expression profiling by next generation sequencing. Project page: https://bitbucket.org/unique379/cbs-mirseq/overview Operating system(s): Linux/Unix. Programming language: Bash, Perl and R. Other requirements: Fastx (v0.0.14), FastQC (v0.10.1), SOLiD_preprocess_filter_v2.pl, cutadapt (v1.6), Bowtie (≤ v1.0), bfast (v0.6.5a), featureCounts (v1.4.6), samtools (v0.1.18), bedtools (v2.23.0), miRspring (v1.2), miRDeep2 (v2.0.05), RNAhybrid (v2.1.1), miRanda (v3.3a), bioconductor packages (> v3). License: GNU GPLv3. Any restriction to use by non-academics: None. *Correspondence: Email: bioinforupesh2009.au@gmail.com; rupesh.kesharwani@jax.org
Current Affiliation Baylor College of Medicine HGSC, Houston TX 77030
Previous Affiliation The Jackson Laborstory of Genomic Medicine 10 discovery Drive Farmington, CT 06032
Previous Affiliation Unit of Immunology and Functional Genomics, Centro Cardiologico Monzino IRCCS, Milan, Italy Tel: (+39) 02 5800 2464; Fax: (+39) 02 5800 2750 Copyright (c) 2019 Kesharwani RK
cite
Rupesh K. Kesharwani, Mattia Chiesa, Riccardo Bellazzi, Gualtiero I. Colombo, CBS-miRSeq: A comprehensive tool for accurate and extensive analyses of microRNA-sequencing data, Computers in Biology and Medicine, Volume 110, 2019, Pages 234-243, ISSN 0010-4825, https://doi.org/10.1016/j.compbiomed.2019.05.019. (https://www.sciencedirect.com/science/article/pii/S001048251930188X)
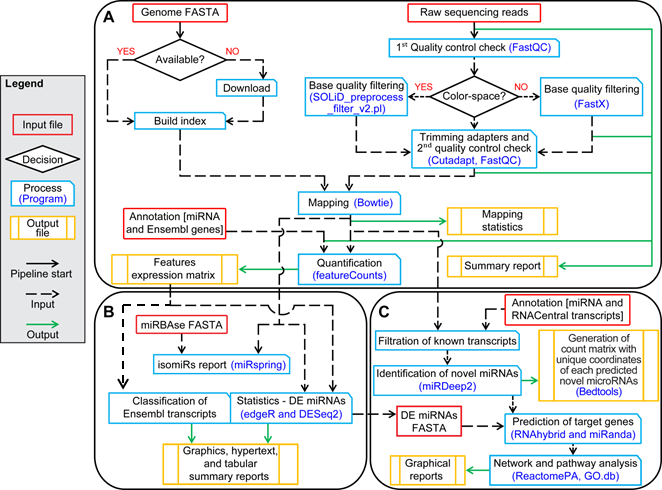
Abstract: Several online and local tools have been developed to analyze microRNA-sequencing (miRNA-Seq) data, but usually they are limited by many factors including: inaccurate processing, lack of optimal parameterization, outdated references plus annotations, restrictions in uploading large datasets, and shortage of biological inferences. In this work, we have developed a fully customized bioinformatics analysis pipeline (Color and Base-Space miRNA-Seq – CBS-miRSeq) for the seamless processing of short-reads miRNA-Seq data. The pipeline has been designed using Bash, Perl, and R scripts. CBS-miRSeq includes modules for read pre- and post-processing (quality assessment, filtering, adapter trimming and mapping) and different types of downstream analyses (identification of miRNA variants (isomiRs), novel miRNA prediction, miRNA:mRNA interaction target prediction, robust differential miRNA analysis, and target gene functional analysis). In this manuscript, we show that re-analysis of two published datasets using the CBS-miRSeq pipeline leads to better performance and efficiency in terms of their pipelines set and biomarker discovery between two biological conditions. Keywords: microRNA; Gene expression profiling; Color-space; Base-space; Bioinformatics pipeline
Installation
================================================================= No installation is required, however pipeline requires prerequisite third party softwares that need to be installed first. However, The simplest way to install CBS-miRSeq on your machine is to download virtual machine image (https://drive.google.com/file/d/0ByG63sGTZ4JTSEVOSlhOVlE1UGs/view?usp=sharing).
Note: For more detail please refer to the CBS-miRSeq source code and manual (https://bitbucket.org/unique379/cbs-mirseq/overview)
Disclaimer: Its rquested to user that each software/tools used within this pipelines, need to be cited properly.
Quick Installation
=================================================================
Building a image from a source of Dockerfile
=================================================================
git clone https://bitbucket.org/unique379/cbs-mirseq.git cd cbs-mirseq docker build . -t cbs-mirseq:v1.0 There should now be a Docker image named cbs-mirseq:v1.0 on your computer.
Verify the successful installation using the command "docker images", which will list every Docker image stored locally on your machine.
Running Docker images:
docker run -ti cbs-mirseq:v1.0
Now, you need to install required R packages using the follwing commands.
Rscript CBS-miRSeq.v1.0/INSTALL/CBS-miRSeq.Required.PackagesV1.1.0.R
Once its completed, You are all set and ready to use CBS-miRSeq.
bash CBS-miRSeq.v1.0/CBS-miRSeq.module1.sh , which will print the help menu.
Start using Source code
$: sudo bash INSTALL/CBS-miRSeq-SystemPackagesInstall.v1.0.sh
Follow the instructions.
Description: This Script will allow user to install system dependencies if missing. Note1: There is no guarantee that every required dependency will install automatically. Note2: Please make certain that the system packages are installed before proceeding with the bioinformatics tool installations. System required necessary packages for Red Hat, Fedora and CentOS (with yum) are: unzip gzip wget git-all automake dos2unix g2-devel perl-CPAN java-1.7.0-openjdk Development Tools libbz2-1.0 zlib1g-devel bzip2-devel libbz2-ocaml libbz2-ocaml-devel libbz2-devel libpng-devel ncurses-devel libncurses5-devel ncurses libncursesw5-devel gcc gcc-gfortran gcc-c++ gd-devel gd-progs libICE-devel libXt-devel make libpdf numpy python-devel python-bzutils python-matplotlib readline-devel zlib-devel
$: bash INSTALL/Install.CBS-miRSeq.dependencies.v1.0.sh
follow the instructions.
Note: Before to run this script; Make sure you have successfully installed miRDeep2 (v2.0.0.5) along with their dependencies. Description: This script aims to install dependencies software and tools required by the CBS-miRSeq pipeline. Note: There is no guarantee that every required dependency will install automatically. Bioinformatics tools that required to execute the Pipeline: Fastx (v0.0.14) FastQC (v0.10.1) SOLiD_preprocess_filter_v2.pl cutadapt (>=v1.6) Bowtie (≤ v1.0) bfast (v0.6.5a) featureCounts (v1.4.6) samtools (v0.1.18) bedtools (v2.23.0) miRspring (v1.2) miRDeep2 (v2.0.05) RNAhybrid (v2.1.1) miRanda (v3.3a)
$: Rscript CBS-miRSeq.v1.0/INSTALL/CBS-miRSeq.Required.PackagesV1.1.0.R Description: This script aims to install R-biocunductor packages required by the CBS-miRSeq pipeline. Note: There is no guarantee that every required package will install automatically. R and Bioconductor packages: RColorBrewer BiocInstaller edgeR DESeq2 EDASeq gplots VennDiagram plotrix ReportingTools hwriter lattice S4Vectors clusterProfiler ReactomePA GO.db biomaRt DOSE networkD3 igraph magrittr stringi KEGGprofile pathview plyr gridExtra grid grDevices XML rJava crayon HTSFilter wordcloud
Quickstart:
=================================================================
Once the system packages, bioinformatics tools and, R are installed, we are ready to run our analysis.
=================================================================
We can execute the pipeline using test data sample (https://drive.google.com/file/d/0ByG63sGTZ4JTUHkxQlJmanV3VW8/view?usp=sharing). (Refer To Test directory)
a) Make a folder where you want to download required results and annotation
$ mkdir Results
$: mkdir annotation
b) Go to the CBS-miRSeq directory
$: cd CBS-miRSeq.v1/Utilities
c) Download the required Annotation files using..
$: bash ./CBS-miRSeq.Annotations.Retrieval.v1.0.sh
Follow the instructions
d) 1. Download the 3' UTR from biomart as instructed in the manual 2. Once you downloaded then execute the script as follow: bash ./CBS-miRSeq.Prepare.UTR.v1.0.sh <Output_dir> <reference_species_3_letter_code> <biomartfasta/mart_export.txt>
############################# e) Start your data analysis- ############################
Please Open the CBS-miRSeq.v1/Input_Info/Module1_Input.txt, Module2_Input.txt, and Module3_Input.txt FILES TO PROVIDE THE REQUIRED INPUTS.
Then RUN the Modules of the Pipeline: ################################################################ Aims of module 1: Download the reference genome; build the index for mapping, Quality control (QC) of the Reads, Trim the 3' adapter, Mapping and Quantification of the features (miRNAs and Ensembl biotypes). Note1: No restrictions regarding the reference organism, user may provide any reference genome. However, this module allow researcher to fetch a few model organism's genome such as Human, Mouse, Rat and Zebrafish. ################################################################
$: cd..
$: bash ./CBS-miRSeq.module1.sh Input_Info/Module1_Input.txt
---> ## Wait till its done.
OPTIONAL STEPS: Note2: In order to save timing to build the index of your reference genome, user may download and build any genome (using bowtie-build) before to run any module and locate the path once module1b ask. In this case user will no longer to use module1 or module1a. Just use module1b for the QC, adapter trimming, Mapping, quantification and report summary of the analysis. ##User need to rename the genome conrresponds to CBS-miRSeq.module1b RENAMING OF THE GENOME: sps.genome_v86.fa ##sps == reference organism in 3 letter code ##v86 is a version release of Ensembl browser. User may fake this if the genome come from different source but make sure header should be like:
chr1 ACGCTGCTGATG chr2 ACGCTGTGTGTCG
#use to build index for base space reads: bowtie-build sps.genome_v86.fa > sps.genome_v86B.fa
#use to build index for color space reads: bowtie-build sps.genome_v86.fa > sps.genome_v86B.fa
#if user got pre-build index from different source, then we recommend to rename it before to use. RENAMING OF INDEX:
for base space reads
sps.genome_v86B.1.ebwt sps.genome_v86B.2.ebwt sps.genome_v86B.3.ebwt sps.genome_v86B.4.ebwt #Reconstruct the genome from index bowtie-build sps.genome_v86B > sps.genome_v86.fa
##for color space reads sps.genome_v86C.1.ebwt sps.genome_v86C.2.ebwt sps.genome_v86C.3.ebwt sps.genome_v86C.4.ebwt
#Reconstruct the genome from index bowtie-build sps.genome_v86C > sps.genome_v86.fa
---->> AS SOON AS, GENOME AND INDEX SET, USER CAN RUN the module1b TO PERFROM ANALYSIS.
################################################################ Aims of module 2: Conducts Diff analysis (DE), Distribution of Ensembl biotype and detection of iso-miRs. Note: Please make sure that groups (along with their replicates) of the expression matrix are set in right place i.e. Control x Treatment (Treatment vs Control). However, it is really depend on your experiments and analysis you wish to perform. i.e. A vs B or B vs A. WARNING!! Output directory cannot be different than module1 (one used in the analysis by module 1). So please make sure it is the same. ################################################################
$: bash ./CBS-miRSeq.module2.sh Input_Info/Module2_Input.txt
---> ## wait till its finish.
################################################################ Aims of module 3: Discovery of Novel miRNA candidates, prediction of target gene of DE miRNAs, Gene enrichment, network and pathway analysis of known and novel miRNAs. WARNING!! Output directory cannot be different than module1 and 2 (one used in the analysis by module 1 and 2). So please make sure it is the same. ################################################################
$: bash ./CBS-miRSeq.module3.sh Input_Info/Module3_Input.txt
---> ## Wait till its finish. ---> Congratulations your analyses being performed.
Note: Example output will create a number of output directories and output inside for further analyses and discoveries.
====================================================================
Optionally, you may execute the sub-modules solitary. It is useful when we want to control the output from every analyses step or to start analysis at different steps of the pipeline.
##Follow the instruction
$: cd /Scripts/CBS-miRSeq.v1/
########################################################## Objective1: User wish to download the reference genome or build the bowtie index (In case user has already downloaded) in order to map their short reads. ##########################################################
$: bash ./CBS-miRSeq.module1a_v1.0.sh
########################################################## Objective2: User to wish to perform Quality Control (QC) of their reads, clip the 3' adapter and mapping the short reads along with expression counts of the map reads. ##########################################################
$: bash ./CBS-miRSeq.module1b_v1.0.sh
################################################################### Objective3: User wish to conduct a Diff Expression analysis between their experimental groups and identification of Ensembl biotype in a sample given. ###################################################################
$: bash ./CBS-miRSeq.module2a_v1.0.sh
######################################################## Objective4: User wish to identify iso-miRs in a sample. ########################################################
$: bash ./CBS-miRSeq.module2b_v1.0.sh
########################################################## Objective5: User wish to Predict Novel miRNAs in a sample. ##########################################################$: bash ./CBS-miRSeq.module3a_v1.0.sh
########################################################## Objective6: User wish to predict the Target gene, perform the gene enrichment, network and pathway analysis of their known and novel miRNAs. ##########################################################
$: bash ./CBS-miRSeq.module3b_v1.0.sh
Please refer the manual for more details and descriptions: CBS-miRSeq.user.manual_v1.pdf
=================================================================
For any assistance and debugging,
Please contact: bioinforupesh2009.au@gmail.com; rupesh.kesharwani@jax.org
Last updated date: 22/05/2019 ##^^^^Thank you for using CBS-miRSeq pipeline^^##