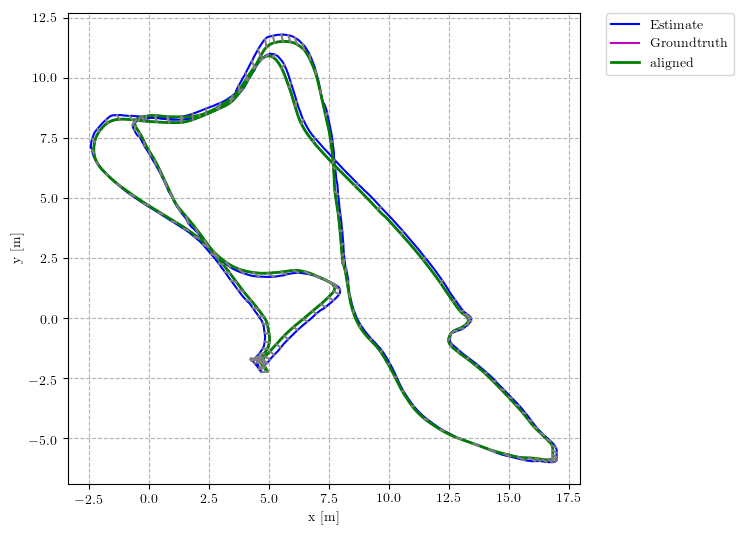
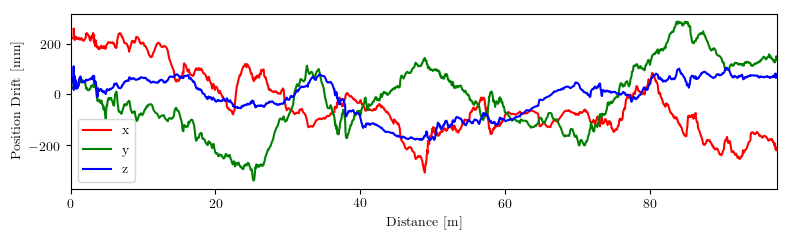
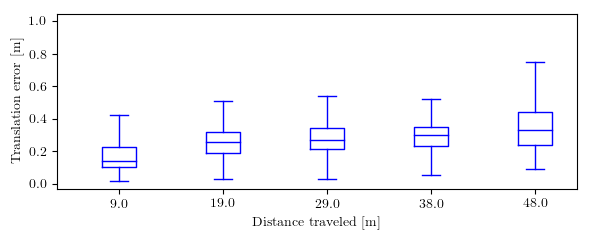
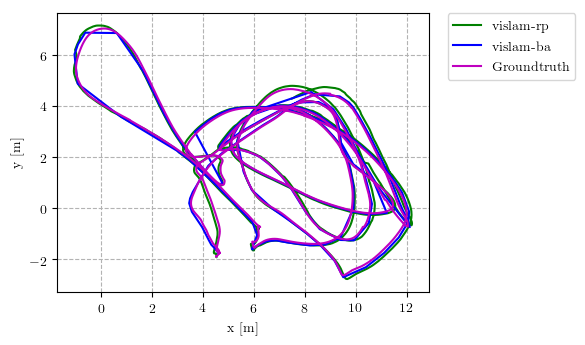
This repository implements common used trajectory evaluation methods for visual(-inertial) odometry. Specifically, it includes
- Different trajectory alignment methods (rigid-body, similarity and yaw-only rotation)
- Commonly used error metrics: Absolute Trajectory Error (ATE) and Relative/Odometry Error (RE)
The relative error is implemented in the same way as in KITTI since it is the most widely used version.
Since trajectory evaluation involves many details, the toolbox is designed for easy use. It can be used to analyze a single trajectory estimate, as well as compare different algorithms on many datasets (e.g., this paper and used in IROS 2019 VIO competition) with one command. The user only needs to provide the groundtruths and estimates of desired format and specify the trajectory alignment method. The toolbox generates (almost) paper-ready plots and tables. In addition, the evaluation can be easily customized.
If you use this code in an academic context, please cite the following paper:
Zichao Zhang, Davide Scaramuzza: A Tutorial on Quantitative Trajectory Evaluation for Visual(-Inertial) Odometry, IEEE/RSJ Int. Conf. Intell. Robot. Syst. (IROS), 2018. PDF
@InProceedings{Zhang18iros,
author = {Zhang, Zichao and Scaramuzza, Davide},
title = {A Tutorial on Quantitative Trajectory Evaluation for Visual(-Inertial) Odometry},
booktitle = {IEEE/RSJ Int. Conf. Intell. Robot. Syst. (IROS)},
year = {2018}
}
The package is written in python and tested in Ubuntu 16.04 and 18.04.
Currently only python2 is supported.
The package can be used as a ROS package as well as a standalone tool.
To use it as a ROS package, simply clone it into your workspace.
It only depends on catkin_simple to build.
Dependencies: You will need install the following:
numpyandmatplotlibfor the analysis/plottingcoloramafor colored console outputruamel.yaml(install) for preserving the order in yaml configurations
Each trajectory estimate (e.g., output of a visual-inertial odometry algorithm) to evaluate is organized as a self-contained folder. Each folder needs to contain at least two text files specifying the groundtruth and estimated poses with timestamps.
stamped_groundtruth.txt: groundtruth poses with timestampsstamped_traj_estimate.txt: estimated poses with timestamps- (optional)
eval_cfg.yaml: specify evaluation parameters - (optional)
start_end_time.yaml: specify the start and end time (in seconds) for analysis.
For analyzing results from N runs, the estimated poses should have suffixes 0 to N-1.
You can see the folders under results for examples.
These files contains all the essential information to reproduce quantitative trajectory evaluation results with the toolbox.
The groundtruth (stamped_groundtruth.txt) and estimated poses (stamped_traj_estimate.txt) are specified in the following format
# timestamp tx ty tz qx qy qz qw
1.403636580013555527e+09 1.258278699999999979e-02 -1.561510199999999963e-03 -4.015300900000000339e-02 -5.131151899999999988e-02 -8.092916900000000080e-01 8.562779200000000248e-04 5.851609599999999523e-01
......
Note that the file is space separated, and the quaternion has the w component at the end.
The timestamps are in the unit of second and used to establish temporal correspondences.
There are some scripts under scripts/dataset_tools to help you convert your data format (EuRoC style, ROS bag) to the above format.
See the corresponding section below for details.
Currently eval_cfg.yaml specifies two parameters for trajectory alignment (used in absolute errors):
align_type:sim3: a similarity transformation (for vision-only monocular case)se3: a rigid body transformation (for vision-only stereo case)posyaw: a translation plus a rotation around gravity (for visual-inertial case)none: do not align the trajectory
align_num_frames: the number of poses (starting from the beginning) that will be used in the trajectory alignment.-1means all poses will be used.
If this file does not exist, trajectory alignment will be done using sim3 and all the poses.
start_end_time.yaml can specify the following (according to groundtruth time):
start_time_sec: only poses after this time will be used for analysisend_time_sec: only poses before this time will be used for analysis
If this file does not exist, analysis be done for all the poses in stamped_traj_estimate.txt.
We can run the evaluation on a single estimate result or for multiple algorithms and datasets.
As a ROS package, run
rosrun rpg_trajectory_evaluation analyze_trajectory_single.py <result_folder>
or as a standalone package, run
python2 analyze_trajectory_single.py <result_folder>
<result_folder> should contain the groundtruth, trajectory estimate and optionally the evaluation configuration as mentioned above.
After the evaluation is done, you will find two folders under <result_folder>:
saved_results/traj_est: text files that contains the statistics of different errorsabsolute_err_statistics_<align_type>_<align_frames>.yaml: the statistics of the absolute error using the specified alignment.relative_error_statistics_<len>.yaml: the statistics of the relative error calculated using the sub-trajectories of length<len>.cached_rel_err.pickle: since the relative error is time consuming to compute, we cache the relative error for different sub-trajectory lengths so that we can directly use them next time.
plots: plots of absolute errors, relative (odometry) errors and the trajectories.
For multiple trials, the result for trial n will have the corresponding suffix, and the statistics of multiple trials will be summarized in files with the name mt_*.
As an example, after executing:
python2 scripts/analyze_trajectory_single.py results/euroc_mono_stereo/laptop/vio_mono/laptop_vio_mono_MH_05
you will find the following in plots:
--recalculate_errors: will remove the error cache file mentioned above and re-calculate everything. Default:False.--png: save plots as png instead of pdf. Default:False--mul_trials: will analyzenruns. In the case ofn > 1, the estimate files should end with a number suffix (e.g.,stamped_traj_estimate0.txt). Default:None
Sometimes, a SLAM algorithm outputs different types of trajectories, such as real-time poses and optimized keyframe poses (e.g., pose graph, bundle adjustment). By specifying the estimation type (at the end of the command line), you can ask the script to analyze different files, for example
--est_type traj_est: analyzestamped_traj_estimate.txt--est_type pose_graph: analyzestamped_pose_graph_estimate.txt--est_type traj_est pose_graph: analyze both. In this case you can find the results in corresponding sub-directories insaved_resultsandplots.
The mapping from the est_type to file names (i.e., stamped_*.txt) is defined in scripts/fn_constants.py. This is also used for analyzing multiple trajectories. You can find an example in results/euroc_vislam_mono for comparing real-time poses and bundle adjustment estimates.
Similar to the case of single trajectory evaluation, for ROS, run
rosrun rpg_trajectory_evaluation analyze_trajectories.py \
euroc_vislam_mono.yaml --output_dir=./results/euroc_vislam_mono --results_dir=./results/euroc_vislam_mono --platform laptop --odometry_error_per_dataset --plot_trajectories --rmse_table --rmse_boxplot --mul_trials=10
otherwise, run
python2 scripts/analyze_trajectories.py \
euroc_vislam_mono.yaml --output_dir=./results/euroc_vislam_mono --results_dir=./results/euroc_vislam_mono --platform laptop --odometry_error_per_dataset --plot_trajectories --rmse_table --rmse_boxplot --mul_trials=10
These commands will look for <platform> folder under results_dir and analyze the algorithms and datasets combinations specified in analyze_trajectories.py, as described below.
The datasets under results are organized as
<platform>
├── <alg1>
│ ├── <platform>_<alg1>_<dataset1>
│ ├── <platform>_<alg1>_<dataset2>
│ └── ......
└── <alg2>
│ ├── <platform>_<alg2>_<dataset1>
│ ├── <platform>_<alg2>_<dataset2>
├── ......
......
Each sub-folder is of the same format as mentioned above.
You need to specify the algorithms and datasets to analyze for the script analyze_trajectories.py.
We use a configuration file under scripts/analyze_trajectories_config to sepcify the details. For example, in euroc_vio_mono_stereo.yaml
Datasets:
MH_01: ---> dataset name
label: MH01 ---> plot label for the dataset
MH_03:
label: MH03
MH_05:
label: MH05
V2_01:
label: V201
V2_02:
label: V202
V2_03:
label: V203
Algorithms:
vio_mono: ---> algorithm name
fn: traj_est ---> estimation type to find the correct file name
label: vio_mono ---> plot label for the algorithm
vio_stereo:
fn: traj_est
label: vio_stereo
RelDistances: [] ---> used to specify the sub-trajectory lengths in the relative error, see below.
RelDistancePercentages: []
will analyze the following folders
├── vio_mono
│ ├── laptop_vio_mono_MH_01
│ ├── laptop_vio_mono_MH_03
│ ├── laptop_vio_mono_MH_05
│ ├── laptop_vio_mono_V2_01
│ ├── laptop_vio_mono_V2_02
│ └── laptop_vio_mono_V2_03
└── vio_stereo
├── laptop_vio_stereo_MH_01
├── laptop_vio_stereo_MH_03
├── laptop_vio_stereo_MH_05
├── laptop_vio_stereo_V2_01
├── laptop_vio_stereo_V2_02
└── laptop_vio_stereo_V2_03
The relative pose error is calculated from subtrajectories of different lengths, which can be specified by the following fields in the configuration file
RelDistances: a set of lengths that will be used for all the datasetsRelDistancePercentages: the lengths will be selected independently as certain percentages of the total length for each dataset. This will be overwritten byRelDistances. If none of the above is specified, the default percentages (seesrc/trajectory.py) will be used.
The evaluation process will generate the saved_results folder in each result folder,
same as the evaluation for single trajectory estimate.
In addition, it will generate plots and text files under results folder comparing different algorithms:
- Under
<platform>_<dataset>_resultsfolder you can find the trajectory top/side views and the boxplots of the relative pose errors on this dataset. all_translation/rotation_rmse.pdf: boxplots of the RMSE of multiple runs for all datasets.<platform>_translation_rmse<algorithm_dataset_string>.txt:Latextable summarizing the RMSE of all configurations.<algorithm_dataset_string>is an identifier generated from the algorithms and datasets evaluated. The values in the table are formated asmean, median (min, max)from the errors of multiple trials.
The tables can be readily used in Latex files.
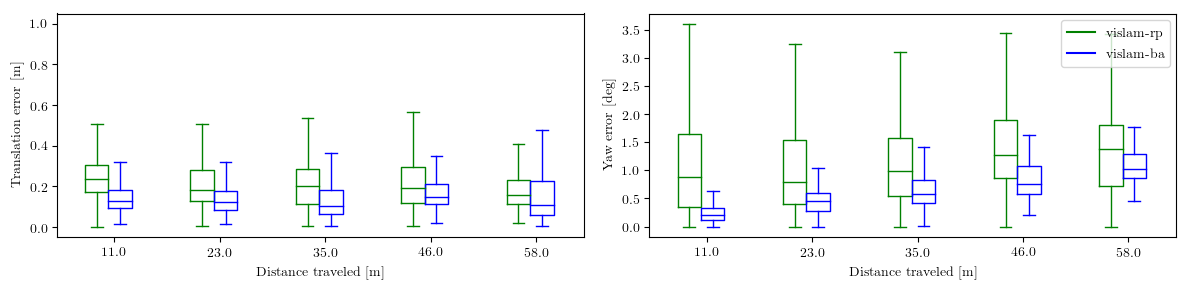
Several example plots (from analyze_trajectories_config/euroc_vislam_mono.yaml) comparing the performance of different algorithms are
Configuration:
configconfiguration file underscripts/analyze_trajectories_config
Paths:
--results_dir: the folder where the<platform>folder will be found. Default:resultsfolder in the toolbox folder.--output_dir: the folder for all the plots and text files. Default:resultsfolder in the toolbox folder.--platform: the folder of results to be found under the<results_dir>. Default:laptop
Analysis options:
-
--mul_trials: how many trials we want to analyze. Default:None. If some algorithm-dataset configuration has less runs, only the available ones will be considered. -
--odometry_error_per_dataset: whether to compute the relative error for each dataset. Default:False. -
--overall_odometry_error: whether to compute and compare the overall odometry error on all datasets for different algorithms. Default:False. -
--rmse_table: whether to generate the table of translation RMSE (absolute error). Default:False.--rmse_table_median_only: per default, the--rmse_tableoption saves median/mean/min/max of multiple runs. Use this option to only save the median.--rmse_boxplot: whether to plot boxplot for RMSE of different datasets (only valid for analysing results from multiple trials). Default:False.
-
--plot_trajectories: whether to plot trajectories. Default:False. By default, many plots are generated, and some of them can be turned off by the following options.--no_plot_side: do not plot side views--no_plot_aligned: do not plot the alignment connection between the groundtruth and estimate--no_plot_traj_per_alg: do not generate plots for each algorithm
Misc:
--recalculate_errors:whether to clear the cache and recalculate everything. Default:False.--png: save plots as png instead of pdf. Default:False--dpi: allow saving at a higher dpi. Default: 300--no_sort_names: do not sort the names of datasets and algorithms (using the order in the configuration file) when plotting/writing the results. Default: names will be sorted.
Under scripts/dataset_tools, we provide several scripts to prepare your dataset for analysis. Specifically:
asl_groundtruth_to_pose.py: convert EuRoC style format to the format used in this toolbox.bag_to_pose.py: extractPoseStamped/PoseWithCovarianceStampedin a ROS bag to the desired format.transform_trajectory.py: transformed a pose file of our format by a given transformation, useful for applying hand-eye calibration to groundtruth/estimate before analysis.
Under scripts, there are also some scripts for conveniece:
recursive_clean_results_dir.py: remove allsaved_resultsdirectories recursively.change_eval_cfg_recursive.py: recursively changing the evaluation parameter within a given result folder.
Most of the error computing is done via the class Trajectory (src/rpg_trajectory_evaluation/trajectory.py).
If you would like to customize your evaluation, you can use this class directly.
The API of this class is quite simple
# init with the result folder.
# You can also specify the subtrajecotry lengths and alignment parameters in the initialization.
traj = Trajectory(result_dir)
# compute the absolute error
traj.compute_absolute_error()
# compute the relative errors at `subtraj_lengths`.
traj.compute_relative_errors(subtraj_lengths)
# compute the relative error at sub-trajectory lengths computed from the whole trajectory length.
traj.compute_relative_errors()
# save the relative error to `cached_rel_err.pickle`
traj.cache_current_error()
# write the error statistics to yaml files
traj.write_errors_to_yaml()
# static method to remove the cached error from a result folder
Trajectory.remove_cached_error(result_dir)After the error is computed, the absolute and relative errors are stored in the trajectory object and can be accessed afterwards:
-
absolute error:
traj.abs_errors. It is a dictionary stores the absolute errors of all poses as well as their statistics. SeeTrajectory.compute_absolute_errorfunction for details. -
relative error:
traj.rel_errors. It is a dictionary stores the relative errors of different distances (using the distance as the key). SeeTrajectory.compute_relative_error_at_subtraj_lenfunction for details.
With the interface, it should be easy to access all the computed errors for customized analysis.
See package.yaml for the list of authors that have contributed to this toolbox.
It might happen that some open-source code is incorporated into the toolbox but we missed the license/copyright information. If you recognize such a situation, please open an issue.
The development of this toolbox was supported by the the Swiss National Science Foundation (SNSF) through the National Centre of Competence in Research (NCCR) Robotics, the SNSF-ERC Starting Grant, and the DARPA FLA program.