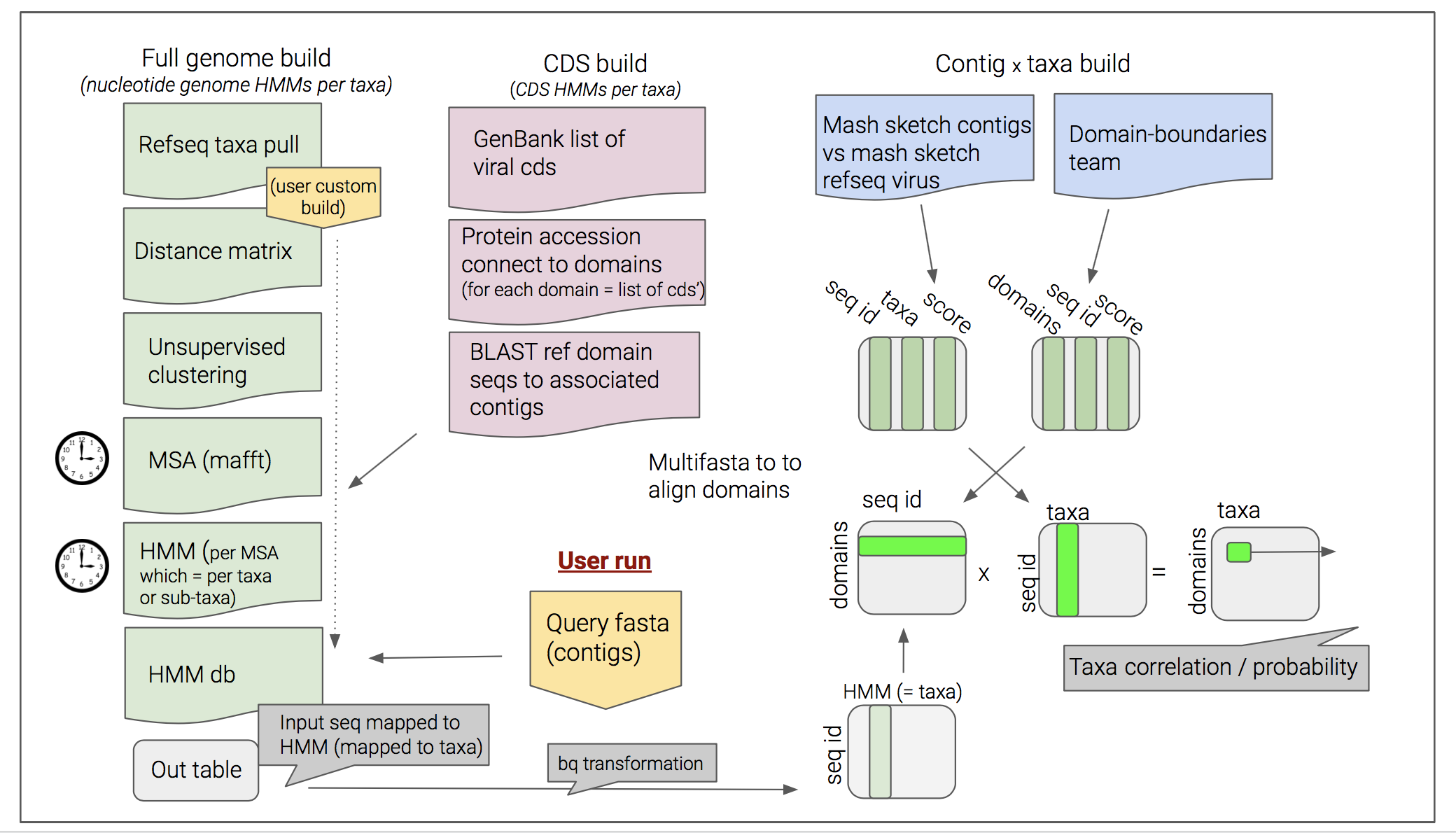
Taxonomy_Domain_Integration
Write MASTER SCRIPT
- Fetch sequences
- Clustering
- Make HMM
- Map models to sequences
- Estimate taxonomy of sequences Mash Define p-value cut-off (-v)
- Combine tables
Generate a distance matrix for Clustering:
Two Wrapper scripts exist for generating distance matrices for unsupervised clustering of sequences. RWrapperA.sh makes use of GNU parallel tools, while RWrapperB.sh does not. Both scripts require a single multifasta file of nucleotide sequences, a matrix output, a names output file, and a PATH.
Usage for parallel implementation:
$ RWrapperA.sh <Seqs.fna> <matrixout> <namesoutput> <PATH>
Usage for non-parallel implementation:
$ RWrapperB.sh <Seqs.fna> <matrixout> <namesoutput> <PATH>
The distance matrix is natively output with rows and columns named by a unique integer which can be mapped to the names text file. This matrix can then be fed into the subsequent clustering approach
Unsupervized clustering for grouping similar genomes:
The distance matrix generated is provided as an input to cluster_similar_fasta_sequences.py along with the a list of names of sequences. An unsupervized clustering approach, based on PCA and K-means, is used to infer the clusters from the distance matrix. The python file also required the actual fasta sequences as an input, through a single multi-sequence fasta, and generates multiple fasta files, each of which contains the fasta sequences identified within a same cluster.
Usage:
$ python3 -d <distance_matrix_file> -n <names_file> -s <Seqs.fna> -o <out_dir>
The fasta files generated have filenames of the form input_files_#.fasta which can then be used directly for whole genome alignment or multiple sequence alignment
Assign taxonomy to the contig sequences
We used Mash to search for the closest viral reference for each contig in the sample. We employed a p-value cutoff (0.05) to limit to top-matching references.
VADIM script
We then linked taxonomic IDs to the identified references, and added SRR accession identifiers to all contigs to make them unique.
python get_taxonomy_id.py ref_viruses_rep_genomes_v5.ids <file with the list files of mash distance>