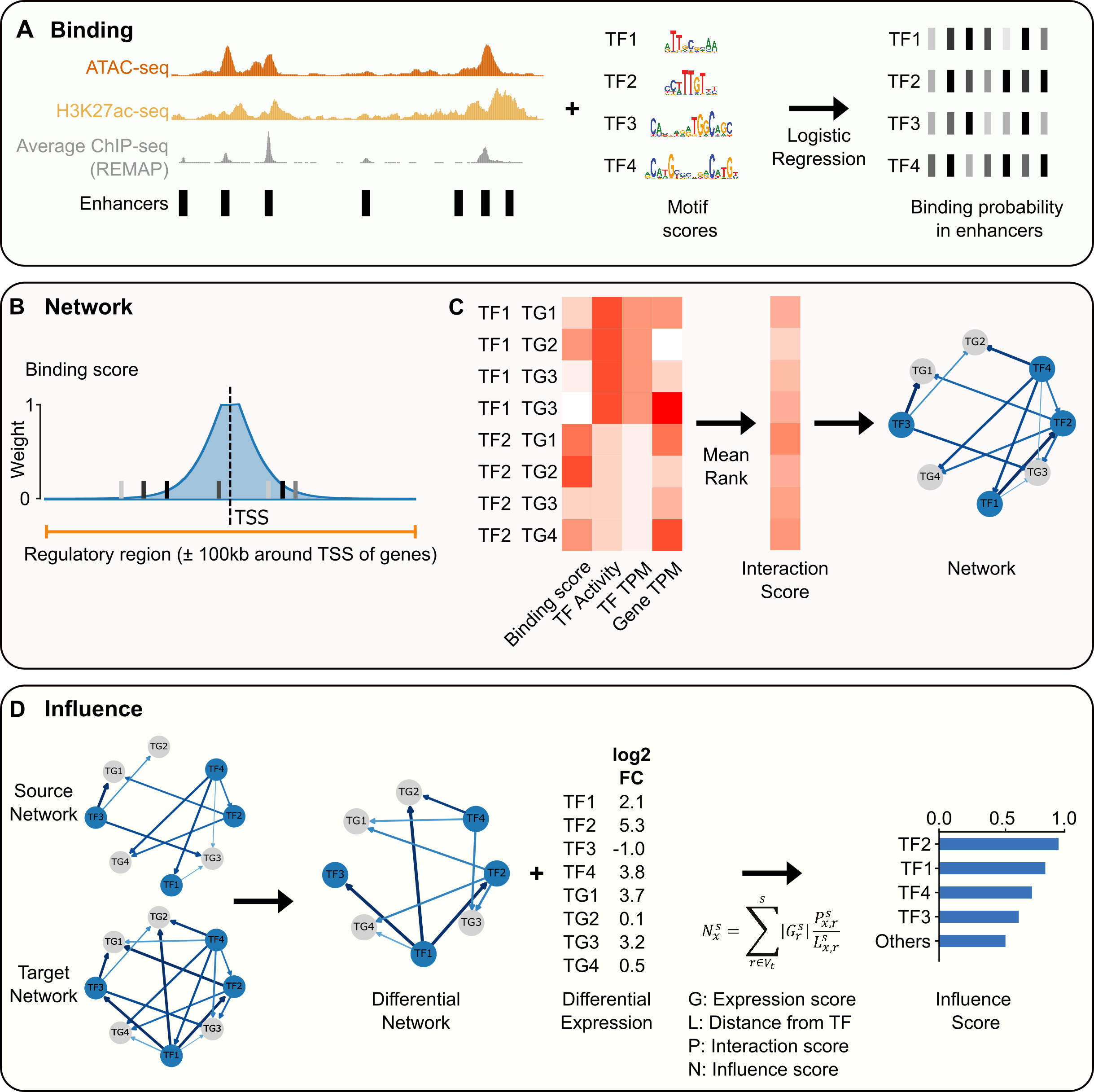
ANANSE is a computational approach to infer enhancer-based gene regulatory networks (GRNs) and to identify key transcription factors between two GRNs. You can use it to study transcription regulation during development and differentiation, or to generate a shortlist of transcription factors for trans-differentiation experiments.
ANANSE is written in Python and comes with a command-line interface that includes 3 main commands: ananse binding, ananse network, and ananse influence. A graphical overview of the tools is shown below.
Check out the ANANSE documentation for
- installation instructions
- command explanations
- input explanations and examples
- usage examples
- FAQ
- and more!
For documentation on the development version see here.
ANANSE: an enhancer network-based computational approach for predicting key transcription factors in cell fate determination Quan Xu, Georgios Georgiou, Siebren Frölich, Maarten van der Sande, Gert Jan C Veenstra, Huiqing Zhou, Simon J van Heeringen Nucleic Acids Research, gkab598, https://doi.org/10.1093/nar/gkab598
scANANSE is a pipeline developed for single-cell RNA-sequencing data and single-cell ATAC-sequencing data. It can export single-cell cluster data from both Seurat or Scanpy objects, and runs the clusters through ANANSE using a snakemake workflow to significantly simplify the process. Afterwards, results can be imported back into your single-cell object.
For more info on this implementation check out the
- scANANSE workflow
- Python package for Scanpy objects
- R package for Seurat objects
- anansnake package for automating multiple ANANSE analyses
- The preferred way to get support is through the Github issues page.
- MIT license
- Copyright 2020 © vanheeringen-lab.