Scripts for the manuscript, Ziyatdinov, et al. "Estimating the effective sample size in association studies of quantitative traits." bioRxiv (2019), https://www.biorxiv.org/content/10.1101/2019.12.15.877217v2.full.
- 336K unrelated individuals of British ancestry
- >600K genotyped SNPs, autosomes, MAF >0.1%, QC
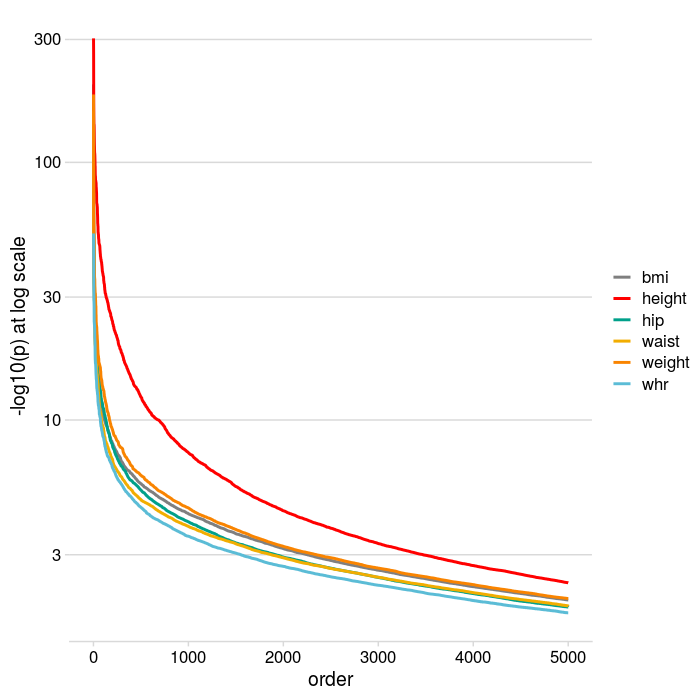
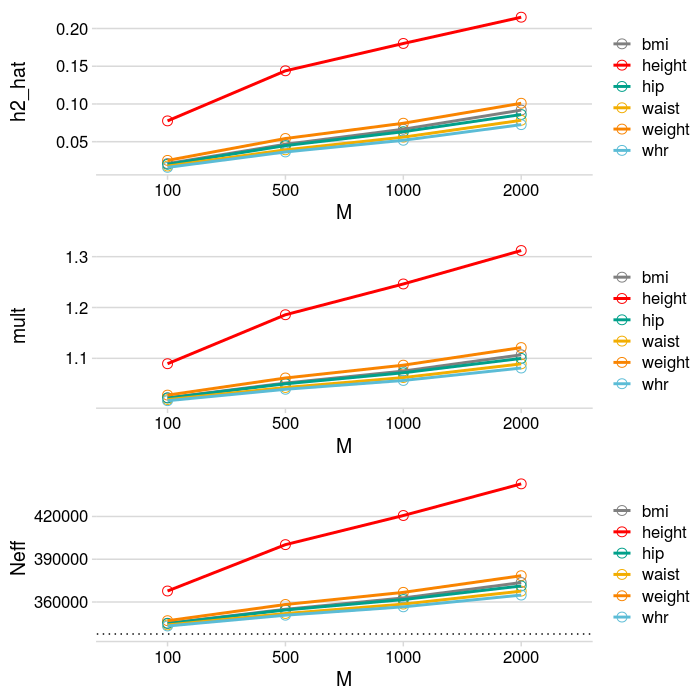
- 6 traits: bmi, weight, waist, hip, height, whr
- impute missing trait values by mean (<1% of missingness)
- project out covariates: age/sex + PC1-20
- apply rank-based inverse normal transformation
Scripts to process raw UK Biobank data are not shared.
- script: scripts/08-gwas-lm-top.R
- snakemake command:
snakemake -s sm.py lm
- plink command:
plink --bfile <bed> --out <out> --clump <assoc> --clump-p1 0.1 --clump-p2 0.1 --clump-r2 0.01 - results: out/clump/
- script: scripts/04-estimate-h2.R
- snakemake command:
snakemake -s sm.py h2 - results: out/h2/
- script: scripts/07-gwas-lmm-loco-top.R
- snakemake command:
snakemake -s sm.py loco