- Dependencies
- On prem environment setup
- Environment setup
- Available workflows
- Write input and submit
- Post run reports
- Create new workflow
- Appendix
- gcloud
- gsutil
- cromwell-tools
- jq
- womtools
Python
- pandas
- jsonschema
Docker images
Set up on prem:
module load wdl
export PYTHONPATH=/gpfs/commons/groups/nygcfaculty/kancero/envs/pip3/wdl-additional/:$PYTHONPATH
Set up:
# Install gcloud cromwell-tools and jq and have them in you path
# Install conda (only need to do this install once)
wget https://repo.continuum.io/miniconda/Miniconda3-latest-Linux-x86_64.sh
bash \
Miniconda3-latest-Linux-x86_64.sh
# Then follow install instructions and then instructions to add conda to your path
# create wdl environment
conda env create -f somatic_dna_wdl/tools/environment.yml
# activate the environment
conda activate wdl
gcloud cromwell-tools jq
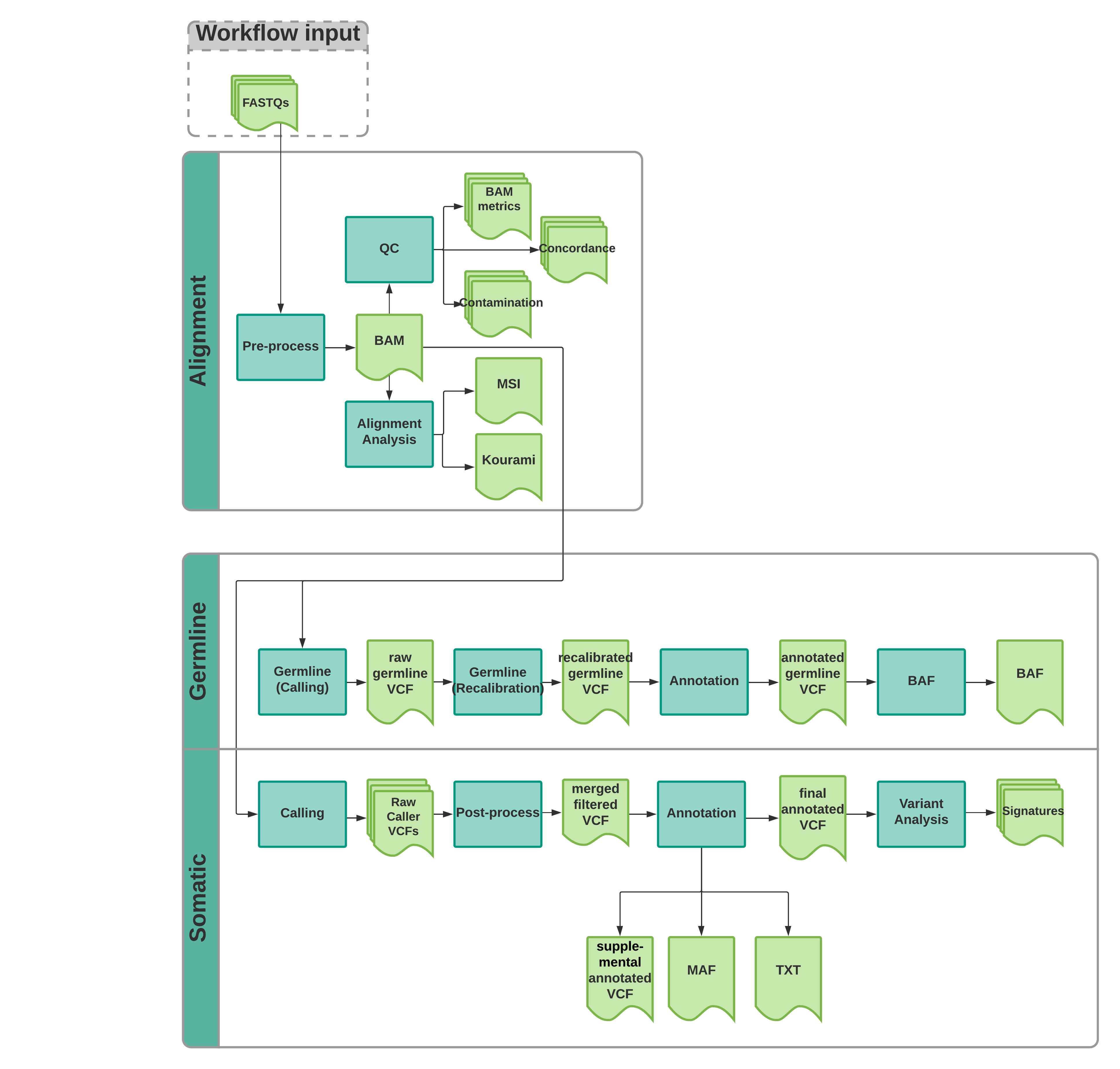
The pipeline is designed to be modular because there are times when we only run a segment and not the entire v7 pipeline. Below are available workflows:
alignment_analysis_wkf.wdl Run Kourami (HLA typing), Mantis (MSI status) and FastNGSAdmix (ancestry) on BAMs
annotate_cnv_sv_wkf.wdl Merge SV calls, filter and annotation both SV and CNV calls
calling_wkf.wdl Run SNV, INDEL, SV and CNV callers on BAMs
filter_intervals.wdl Prep reference files by filtering using a BED file of intervals
gdc_wkf.wdl Compare VCF files
germline_wkf.wdl Call germline SNVs and INDELs and output a filtered and unfiltered
annotated VCF
kourami_wkf.wdl Run Kourami (HLA typing)
merge_vcf_wkf.wdl Merge, filter and annotate v7 pipeline calls
report_mini_wkf.wdl In dev report writing workflow
report_wkf.wdl In dev full report writing workflow
somatic_bam_wkf.wdl Full v7 pipeline starting from BAMs: SNV, INDEL, SV and CNV calling
filtering and annotating, germline calls and BAF (HaplotypeCaller),
DeconstructSigs (Mutational Signatures), contamination and concordance
(Conpair), Kourami (HLA typing), Mantis (MSI status) and
FastNGSAdmix (ancestry)
somatic_wkf.wdl Full v7 pipeline starting from FASTQs: Alignment, QC, SNV, INDEL, SV
and CNV calling filtering and annotating, germline calls and BAF
(HaplotypeCaller), DeconstructSigs (Mutational Signatures),
contamination and concordance (Conpair), Kourami (HLA typing), Mantis (MSI status),
and FastNGSAdmix (ancestry)
tests_wkf.wdl Automated comparison of prior pipeline run to current pipeline run output.
variant_analysis_wkf.wdl Run DeconstructSigs (Mutational Signatures)
wdl_structs.wdl Custom Struct objects to reference primary and secondary files together
or group ids and related files
Script somatic_dna_wdl/run.sh will first quickly validate the WDL workflow. Next it will use the WDL
to determine which variables are required. All required variables will be defined from the
Reference JSON in config, the pairing/sample info or the custom inputs JSON.
Most URIs for files will be validated to ensure you can read the file (unless the --skip-validate flag is used). If a pipeline has the variable production and/or external and these are set to true then the pipeline will skip tasks that require private NYGC files or licenses. Because of this if either production or external are true for a workflow private NYGC files will not have their uris validated.
Next, the input JSON will be compared to the original WDL and the program will exit if any required variable could not be found and will need to be provided in the custom input JSON.
Then the workflow will be submitted to the cromwell server.
In addition to submitting the command, this will create an output file that you should save. It contains information about the project, pipeline version, cromwell options, inputs. It will also contain the workflow UUID. This will be used after the run to agregate information about the run.
somatic_dna_wdl/run.sh -h
run.sh [-h] --options OPTIONS --wdl-file WDL_FILE
--url URL --log-dir LOG_DIR
--project-name PROJECT_NAME
[--library {WGS,Exome}]
[--genome {Human_GRCh38_full_analysis_set_plus_decoy_hla, Human_GRCh38_tcga}]
[--pairs-file PAIRS_FILE]
[--samples-file SAMPLES_FILE]
[--interval-list {SureSelect_V6plusCOSMIC.target.GRCh38_full_analysis_set_plus_decoy_hla}]
[--custom-inputs [CUSTOM_INPUTS [CUSTOM_INPUTS ...]]]
[--skip-validate]
[--dry-run]
DESCRIPTION: validate workflow, create input json and submit workflow to cromwell.
Script requires jq, cromwell-tools, gcloud to be in the path.
Script shows submission command and command to check status
in the STDERR stream.
Creation of input JSON:
The WDL is used to determine which variables are required.
Required or optional variables are defined from custom inputs JSON.
Any required variable not defined in the custom inputs JSON will be defined from the
reference JSONs in the config directory (as long as the variable names are identical).
The pairing/sample info CSVs (--pairs-file/--samples-file) are used to create pairRelationships and
(if columns named tumorBam and normalBam exist) map BAMs to pairs.
If "production" or "external" inputs are true then validation of NYGC internal-only files is skipped
The pipelines are written to skip tasks that localize these files if "production" or "external" are true
so any inability to read these files will not negatively affect the run.
-h, --help show this help message and exit
--url URL Cromwell server URL (required)
--log-dir LOG_DIR Output directory for all logs and reports
related to this workflow UUID (required)
--options OPTIONS Options json file for cromwell (required)
--wdl-file WDL_FILE WDL workflow. An input JSON that matches this
WDL workflow will be created (required)
--file-out FILE_OUT Output file for project info
--custom-inputs-out CUSTOM_INPUTS_OUT
Output file for custom workflow inputs
--custom-labels-out CUSTOM_LABELS_OUT
Output file for workflow labels
--library {WGS,Exome}
Sequence library type.
--genome {Human_GRCh38_full_analysis_set_plus_decoy_hla, Human_GRCh38_tcga}
Genome key to use for pipeline.
--project-name PROJECT Project name associated with account.
--pairs-file PAIRS_FILE
CSV file with items that are required to have
"tumor", "normal" and "pairId" in the columns.
Optionally, include "tumorBam", "normalBam" columns to create
"pairInfos" and "normalSampleBamInfos" automatically.
--samples-file [SAMPLES_FILE]
Not generally required. If tasks only require
sampleId and do not use pairing information sample
info can be populated with a CSV file. The CSV file
requires a columns named ["sampleId"].
--interval-list {SureSelect_V6plusCOSMIC.target.GRCh38_full_analysis_set_plus_decoy_hla}
File basename for interval list.If not supplied the
default (the SureSelect interval list for your genome)
will be used (only needed for future Exome workflows)
--custom-inputs [CUSTOM_INPUTS]
Optional JSON file with custom input variables. The
name of the variable in the input file must match the
name of the variable in the WDL workflow. It is not
required that the input specify the workflow. By
default the input will be added to the top-level
workflow. Any variable defined in in this JSON will
overwrite any reference variable in the the config
directory or workflow default.
--skip-validate Skip the step where input files are validated.
Otherwise all gs//: URIs will be checked to see that a
file exists. Disable with caution. Cromwell will launch
instances and run without checking. Test a small pairs
file to ensure all references exist and at least some
sample input files can be read by the current user.
--dry-run Skip the step where the job is submitted to cromwell-tools.
Command
# Create input json
cd ${working-dir}
../somatic_dna_wdl/run.sh \
--log-dir ${working-dir} \
--url ${url} \
--project-name ${lab_quote_number} \
--pairs-file ${tumor_normal_pairs_csv} \
--library WGS \
--genome Human_GRCh38_full_analysis_set_plus_decoy_hla \
--wdl-file somatic_dna_wdl/somatic_wkf.wdl \
--options options.json \
--custom-inputs-out ${lab_quote_number}.inputs.json \
--custom-labels-out ${lab_quote_number}.labels.json \
--file-out ${lab_quote_number}.projectInfo.json
somatic_wkfInput.json- inputs for cromwell${lab_quote_number}_projectInfo.json- contains project info like the current list of samples/pairs and the library type as well as the pipeline version (tag and commit).${lab_quote_number}_project.<DATE>.RunInfo.json- contains projectInfo, workflow Input, workflow UUID.
Use the cromwell-tools status command printed to the screen when you submitted the workflow. Alternately, lookup the workflow UUID in the lab-number_project.<DATE>.RunInfo.json and run:
cromwell-tools status \
--url ${url} \
--username $(gcloud secrets versions access latest --secret="cromwell_username") \
--password $(gcloud secrets versions access latest --secret="cromwell_password") \
--uuid ${uuid}
After workflow finishes and the status is SUCCEEDED or while it is running use run_summary.sh to review run metrics and cost:
USAGE: run_summary.sh -u URL -n PROJECT_NAME -g GCP_PROJECT -d log_dir [-r RUNINFO_JSON] [-i UUID] [-b BILLING EXPORT] [--from-billing] [-p PAIRS_FILE] [-s SAMPLES_FILE]
DESCRIPTION: monitor or summarize cromwell workflow.
Script requires jq to be in the path.
If no *.RunInfo.json file OR uuid is provided then
the most recent file in the log dir will be used
Script generates summary of any workflow.
-h, --help Show this help message and exit
--billing-export BILLING The name of the table for the SQL query
of the billing table.
--from-billing Skip regenerating runtime metrics. Use existing
metrics files; add the cost values from the billing
table; and plot the results (including billing).
--gcp-project GCP_PROJECT GCP project id for api queries.
--run-info-json RUNINFO_JSON Optional file that includes any sampleIds and the
main workflow UUID.
--url URL Cromwell server URL (required)
--log-dir LOG_DIR Output directory for all logs and reports
related to this workflow UUID (required)
--project-name PROJECT_NAME Project name associated with account.
--pairs-file PAIRS_FILE
Optional, CSV file with items that are required to have
"tumor", "normal" and "pairId" in the columns.
Optionally, include "tumorBam", "normalBam" columns to create
"pairInfos" and "normalSampleBamInfos" automatically.
--samples-file [SAMPLES_FILE]
Not generally required. If tasks only require
sampleId and do not use pairing information sample
info can be populated with a CSV file. The CSV file
requires a columns named ["sampleId"].
Command:
bash templates/run_summary.sh \
-u ${url} \
-d ${log_dir} \
-p ${gcp_project} \
-n ${project_name}
Optionally add -r ${run_info} or --uuid ${uuid} to the command. Otherwise, the script will use the
most recent *RunInfo.json file in the ${log_dir}.
If you do not have a *RunInfo.json you can start with the workflow uuid
(a sample or pair csv file can also be indicated but is not required).
bash \
templates/run_summary.sh \
-u ${url} \
-d ${log_dir} \
-b ${billing_export} \
-n ${project_name} \
-g ${gcp_project} \
--uuid ${uuid} \
--samples-file ${sample_id_list}
is the name of the table for the SQL query) to add cost per instance_id to results:
-b ${billing_export}
If the workflow finishes and the status is SUCCEEDED and you have run run_summary.sh. It will output several log files:
${lab_quote_number}<WORKFLOW_UUID>_outputInfo.json :
Conatins relevant information on the run including:
named_files: list of output files (in the final location)outputs: map between workflow output object-name and object (in the final bucket/location)options: options values from runworkflow_uuid: uuidrun_date: run_datestatus: statuspair_association: map of pair_ids to theoutputsmap for just that pair (searches for the pair_id followed by . _ or / )sample_association: map of sample_ids to theoutputsmap for just that sample (searches for the sample_id followed by . _ or / )
Note: Pair association only works if the pair_id is used in the filename followed by a ., / or a _. Sample association only works if the sample_id is used in the filename followed by a . or a _.
${lab_quote_number}<WORKFLOW_UUID>_outputMetrics.csv
the file includes the following metrics calculated from the BigQuery cromwell monitor:
'task_call_name', 'wdl_task_name', 'sub_workflow_name', 'workflow_name',
'execution_status', 'cpu_count', 'disk_types', 'docker_image',
'attempt', 'backend_status', 'preemptible', 'instance_name', 'zone',
'project_id', 'return_code', 'workflow_id', 'shard', 'start_time',
'end_time', 'localization_m', 'inputs', 'main_workflow_name', 'labels',
'id', 'instance_id', 'cpu_platform', 'mem_total_gb', 'disk_mounts',
'disk_total_gb', 'actual_start_time', 'wait_time_m', 'actual_runtime_m',
'max_cpu_used_percent', 'max_disk_used_gb', 'max_mem_used_gb',
'run_time', 'run_time_m', 'cpu_time_m', 'sample_task_run_time_h',
'sample_task_core_h', 'sample_subworkflow_core_h',
'sample_subworkflow_run_time_h', 'subworkflow_max_mem_g',
'sample_workflow_core_h', 'sample_workflow_run_time_h',
'workflow_max_mem_g', 'main_workflow_id', 'cromwell_workflow_id',
'avg_capacity_cost', 'avg_core_cost', 'avg_egress_cost', 'avg_ram_cost',
'avg_total_cost', 'runtime_scaled_total_cost', 'machine_type',
'disk_type'
${lab_quote_number}<WORKFLOW_UUID>_outputMetrics.html
This file includes plots of runtime metrics.
${lab_quote_number}<WORKFLOW_UUID>_outputCosts.csv
This is the table returned from the billing db on the cloud (only created if the billing table flag is used)
${lab_quote_number}<WORKFLOW_UUID>.outputMetrics.cost.csv
This is the metrics table with the costs added (only created if the billing table flag is used)
${lab_quote_number}<WORKFLOW_UUID>.outputMetrics.total.csv
This is the end-to-end cost for the pipeline (according to the billing table) (only created if the billing table flag is used)
Use a style guide to write you WDL files.
- Write a new workflow (e.g.
wdl_port/new_wkf.wdl) using structs fromwdl_structs.wdlwere needed. Use the variable fromconfig/fasta_references.jsonandconfig/interval_references.jsonin your workflow (e.g.referenceFa)
- keep your tasks and sub workflow in a separate WDL file in a subdirectory. That workflow should run a section of the pipeline on one sample/pair.
- In the main directory make a WDL that runs the sub workflow(s) for a list of
sampleInfoorpairInfoobjects. - Alternately add your subworkflow to an existing main workflow (e.g.
calling_wkf.wdl)
- Add any new required resource files to
config/fasta_references.jsonandconfig/interval_references.json. - Upload any new resource files:
# modified or in-house files
gsutil cp \
${file} \
gs://${resources_project}/GRCh38_full_analysis_set_plus_decoy_hla/internal/
# external reference files
gsutil cp \
${file} \
gs://${resources_project}/GRCh38_full_analysis_set_plus_decoy_hla/external/
- Confirm all workflows are still valid and commit changes
Also confirm that all workflows continue to be valid before commiting your changes
for file in */*wdl *wdl; do
echo $file;
womtool validate $file;
done
# commit your changes and create new inputs file (with new branch and commit info)
git commit -m "feat: my new workflow"
- Run the new workflow as before
Run the following once to generate a default credentials file
$ gcloud auth application-default login
See for more details: https://google-auth.readthedocs.io/en/latest/reference/google.auth.html#google.auth.default.
- biqsec2 norm
- svaba?
- lancet
- mantis kmer counter ?
- strelka2
- mutect2
- gridss
- manta
- all samtools/pysam steps
- kourami w/ custom prep
We are in the process of setting up a public issue tracker. In the mean time please send suggestions, questions or issues to jshelton@nygenome.org
New tag releases are made for any new/changed step in the main alignment, calling, merging, annotation, or addtional analysis (e.g. MSI, HLA, etc) pipeline.
7.4.1 Minor adjustments:
- Minor changes to resource requests and naming conventions
7.4.0 Log4j fix, remove SvABA, 1000g PON:
- Ability to skip coverage check for a sample.
- Add bam-to-cram conversion.
- Remove Svaba as SV caller
- Switch to SSD for callers
- Add WGS PON from 1000g deep coverage samples
- Update Gatk and Gridss versions to avoid log4j security bug
7.3.10 Ancestry pipeline:
- add fastngsadmix ancestry analysis as standalone workflow and to somatic_bam_wkf
- add ancestry to somatic_wkf.wdl
7.3.5 Fix for bug in Bioconductor and Gridss:
- fix: update gridss workflow to skip non-canonical chrome that can cause failures
- fix: update gridss docker image for version with fix for new bug in Bioconductor
- fix: skip using private files for disk size estimate
7.3.3 Refactor:
- switch from tags to sha has for docker images
- make bicseq2 config files reference files
- begin pipeline output docs
- switch to merge specific preMergedPairVcfInfo object for merge
- add environment.yml
- get rc file for more complete uuid list (for any not listed in subworkflow final output)
- added SNV/INDEL only pipeline
- added SNV/INDEL, SV and CNV only pipeline
- added 'external' boolean to skip internal files
- switch to public docker images
- switch to public reference files
- add input examples and example commands in docs
- remove --read-length flag and replace with reading from input json
- remove all private files from input JSON
7.3.2 Refactor:
- adjust mem and disk size
- finalize DeconstructSigs workflow
- add highMem flag
- calculate jvmHeap from mem
- decrease mem and disk sizes
- switch from local-disk SSD/HDD to local-disk LOCAL for steps that run on bams
- add gridss arrange steps that works with cache
- update resource usage scripts
7.3.1 Refactor:
- add deconstructsigs
- get chr6 coordinates from smaller file
- adjust threads
- speed up allele counts (chrom splits)
7.2.0 GDC references:
- add Human_GRCh38_tcga
- populate BAMs from a table
- restrict to .bai index extensions
- add workflow to reheader interval lists for external reference files
We are in the process of setting up a public issue tracker. In the mean time please send suggestions, questions or issues to jshelton@nygenome.org