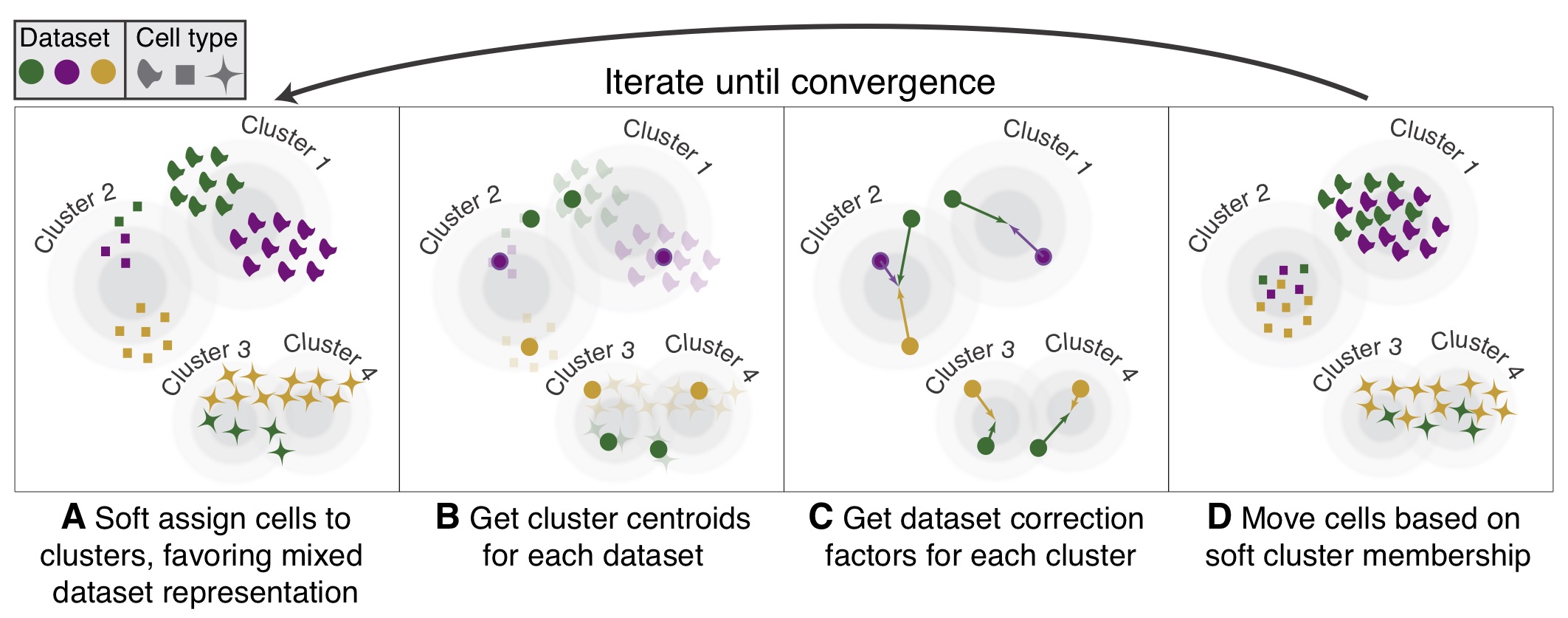
Check out the latest preprint of Harmony on bioRxiv
To run Harmony, open R and install directly from github using the following commands:
library(devtools)
install_github("immunogenomics/harmony")
Try out Harmony on your single cell dataset! If you already have PCA embeddings for your cells and a vector that defines some batch you want to integrate over, Harmony will do the rest:
library(harmony)
my_harmony_embeddings <- HarmonyMatrix(my_pca_embeddings, my_batch_vector)
Now your cell embeddings will be less dependent on your batch variable. Do you want to align the data even more? Consider increasing the alignment parameter:
my_harmony_embeddings <- HarmonyMatrix(my_pca_embeddings, my_batch_vector, theta = 4)
If you already have a Seurat workflow for analyzing your single cell data, check out the tutorial below. You'll only need to make two changes to your code:
- Run Harmony with the
RunHarmonyfunction - In downstream analyses, use the Harmony embeddings instead of PCA. For instance, in RunTSNE, specify
reduction.use = "harmony", or in FindClusters, specifyreduction.type = "harmony".
This vignette is based on the original in Seurat
Coming soon!
This section will show you how to align cells from donors and tissues at the same time.