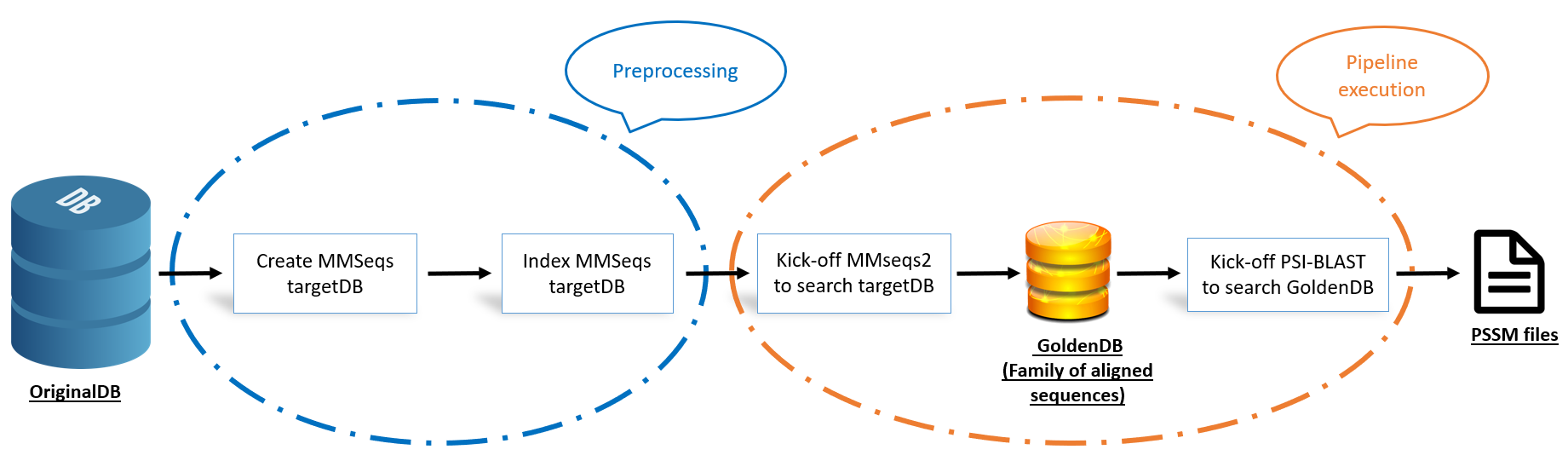
EPSAPG is a pipeline combining modules from state-of-the-art methods to quickly generate extensive protein sequence alignment profiles. It is a fast alignment pipeline that runs the muscles of MMseqs2 and generates PSI-BLAST output (profile and pssm).
I started researching this project idea back during my stay as an AI researcher at Rostlab, Technical University of Munich.
Main entry point of the software is "epsapg.cpp".
If you use EPSAPG in your work, please cite the following publication:
- I. Arab, EPSAPG: A Pipeline Combining MMseqs2 and PSI-BLAST to Quickly Generate Extensive Protein Sequence Alignment Profiles, In 2023 IEEE/ACM 10th International Conference on Big Data Computing, Applications and Technologies (BDCAT ’23), December 4–7, 2023, Taormina, Messina, Italy. ACM, New York, NY, USA, 9 pages. doi:10.1145/3632366.3632384
❗ Before you start compiling any code, make sure you have a machine that supports SSE4.1 or AVX2. You can check that by typing the following commands
cat /proc/cpuinfo | grep -c sse4_1 > /dev/null && echo "SSE4.1 Supported" || echo "SSE4.1 Unsupported"
OR
cat /proc/cpuinfo | grep -c avx2 > /dev/null && echo "AVX2 Supported" || echo "AVX2 Unsupported"
Once the hardware requirements are met, you can move forward to the software requirements.
❗ Compiling the following software will require 'git', 'g++' (7.3 or higher) and 'cmake' (3.0 or higher).
The commands bellow are taillored to CentOS 7, a Linux distribution. You can lookup the equivalent command for each step. In most of the time, you just need to replace 'yum' by 'apt-get'. Refer to the support page bellow to figure out the equivalent in other Linux distributions: https://help.ubuntu.com/community/SwitchingToUbuntu/FromLinux/RedHatEnterpriseLinuxAndFedora
You can either install gcc from repository by typing:
yum -y install gcc
In order to include c++ library as well, you may want to install gcc-c++. This package will compile files with extensions that indicate they are C source as C++, instead of as C. You can install it by typing:
yum -y install gcc-c++
gcc --version
Depending on your version of your CentOS, you may see that GCC is not the last version available. In my machine, CentOS 7, the version distributed in this OS is GCC 4.8.5
Since we need GCC 7.3 or higher, we may need to install it from source. To do so, follow the next steps:
Install GCC from source by typing
wget http://ftp.mirrorservice.org/sites/sourceware.org/pub/gcc/releases/gcc-7.3.0/gcc-7.3.0.tar.gz
tar zxf gcc-7.3.0.tar.gz
cd gcc-7.3.0
Then, you have to install bzip2 and run the ‘download_prerequisites’ script from the top level of the GCC source tree to download some prerequisites needed by GCC:
yum -y install bzip2
./contrib/download_prerequisites
After all the prerequisites are installed and in order to configure the GCC build environment, run the following 'configure' script:
./configure --disable-multilib --enable-languages=c,c++
Then at the end run the following command to compile the source code. It is recommanded to start a screen before you run them since the compilation may take more than an hour
screen -S gcc
make -j 4
make install
Boost is a set of libraries for the C++ programming language that provide support for tasks and structures such as linear algebra, pseudorandom number generation, multithreading, image processing, regular expressions, and unit testing. EPSAPG make use of different optimized algorithms available in Boost, namely tokenizer, string algorithms, and uuid library. To install the Boost, type the following:
yum install boost-devel
MMseqs2 (Many-against-Many sequence searching) is a software suite to search and cluster huge proteins/nucleotide sequence sets. It is recommanded to compile MMseqs2 from source as it has the advantage to be optimized to the specific system, which should improve its performance.
In order to compile from source, clone the repo from GitHub and run the following commands (coppied from the GitHub README file)
git clone https://github.com/soedinglab/MMseqs2.git
cd MMseqs2
mkdir build
cd build
cmake -DCMAKE_BUILD_TYPE=RELEASE -DCMAKE_INSTALL_PREFIX=. ..
make
make install
export PATH=$(pwd)/bin/:$PATH
Now you have to build the mmseqsDB of your fasta database and then compile an index table for it in order to acheive faster runs later. For more details about the project and the commands to use for building the DB and index table, refer to the repository in GitHub:
https://github.com/soedinglab/MMseqs2
BLAST is the Basic Local Alignment Search Tool. It uses an index to rapdily search large sequence databases. Please refer to the following liking to install it:
https://angus.readthedocs.io/en/2019/running-command-line-blast.html#what-is-blast
To compile EPSAPG, clone the repo then run 'make'. After succesful compilation, the executable will be placed in the bin folder. After that include the path into the system variable:
git clone https://github.com/git@github.com:issararab/EPSAPG.git
cd EPSAPG
make
export PATH=$(pwd)/bin/:$PATH
| Parameter | Mandatory/Optional | Input | Description |
|---|---|---|---|
| -query | Mandatory | Fasta_File_In | Input file name in fasta format |
| -interm_path | Optional | folder_path | If run with indextable, which is the recommanded option, MMseqs2 stores intermediate results in tmp folder. (If not specified, the default path is 'tmp' folder in the same location as targetDB. Same one used while indexing the DB.) |
| -db_load_mode | Optional | int_value[1/2] | Option for using instant search power of MMseqs2, set to 2. (default 1, loads db from disk). This option enhaces the search unless the indextable is loaded and locked into memory. Refer to: How to search small query sets fast in the MMseqs2 documentation. |
| -dbsize | Optional | size | Effective length of the database. Default uses the amino acid length of MMseqs2 output result. For accurate statistics of the profile, provide the main database length. |
| -use_sw_tback | Optional | boolean[0/1] | Compute locally optimal Smith-Waterman alignments(default is set to 0) |
| -max_seqs | Optional | int_value | maximum result sequences per query (this parameter affects the sensitivity). Default is set to 1000. |
| -threads | Optional | int_value | number of cores used for the computation (uses all cores of the machine by default) |
| -output_profile | Optional | boolean[0/1] | Enable or disable outputing alignment profile of a query sequence against a database. If disabled, it will print on console.(enabled by default) |
| -output_pssm | Optional | boolean[0/1] | Enable or disable outputing position-specific scoring matrix checkpoint file (disabled by default) |
| -output_ascii_pssm | Optional | boolean[0/1] | Enable or disable outputing ASCII version of position-specific scoring matrix checkpoint file (disabled by default) |
❗ For running the pipeline succesfully, make sure first to create MMseqs database and index table of your main DB with the name targetDB. The pipeline identifies targetDB as the database to search on. The following are the three main commands to search with EPSAPG (read details in MMseqs documentation cited above):
#Compile a MMseqs2 database
mmseqs createdb main/DB.fasta targetDB
# Compile an index table of the DB
mmseqs createindex targetDB tmp
#Launch the search
epsapg -query query/example.fasta
# alternative precise intermediate folder path
mmseqs createindex targetDB /path/tmp
epsapg -query query/example.fasta -interm_path /path/tmp
Note: MMseqs database and indextable are compiled only once, as long as the main database did not change. Refer to "EPSAPG Command paramters" section for more search options.
Enjoy your super fast alignments :)