microbiomer
The goal of microbiomer is to provide a small set of tools to more
seamlessly integrate tools/functions from both the
phyloseq and
tidyverse-packages.
Note this is a package in development; although the functions were not yet tested extensively within the context of this package, they were tested across several projects as source code. Nevertheless, use these functions at your own risk and please file a report if you come across issues.
Installation
You can install the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("wsteenhu/microbiomer")The package has not been submitted to CRAN.
Example
Load packages
library(microbiomer); library(phyloseq)
library(tidyverse)
#> ── Attaching packages ─────────────────────────────────────── tidyverse 1.3.0 ──
#> ✓ ggplot2 3.3.3 ✓ purrr 0.3.4
#> ✓ tibble 3.0.6 ✓ dplyr 1.0.4
#> ✓ tidyr 1.1.2 ✓ stringr 1.4.0
#> ✓ readr 1.3.1 ✓ forcats 0.5.1
#> ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
#> x dplyr::filter() masks stats::filter()
#> x dplyr::lag() masks stats::lag()Load data
data(ps_NP)
ps_NP
#> phyloseq-class experiment-level object
#> otu_table() OTU Table: [ 1793 taxa and 287 samples ]
#> sample_data() Sample Data: [ 287 samples by 5 sample variables ]
#> tax_table() Taxonomy Table: [ 1793 taxa by 8 taxonomic ranks ]The package includes nasopharyngeal microbiota data collected at 1 week,
1 month and 1 year of life. For more information, see ?ps_NP.
Convert
ps_NP_RA <- otu_table(ps_NP) %>%
to_RA()
ps_NP_RA[1:5,1:3] # depict example
#> OTU Table: [5 taxa and 3 samples]
#> taxa are rows
#> 121142010012 121144010012 121150010012
#> Moraxella_2 0.0002340276 0.0002837819 0.6826699800
#> Corynebacterium_5 0.0000000000 0.1827082249 0.0743064326
#> Dolosigranulum_pigrum_7 0.3214870783 0.2691671002 0.2292948222
#> Staphylococcus_3 0.0322958109 0.0283308897 0.0001018454
#> Haemophilus_9 0.0000000000 0.0004256728 0.0000000000to_RA converts the otu_table-object nested within a phyloseq-object
into a total-sum-scaled table (relative abundances instead of raw
reads).
Filter
ps_NP %>% ntaxa()
#> [1] 1793
ps_NP %>%
pres_abund_filter()
#> A total of 224 ASVs were found to be present at or above a level of confident detection (0.1% relative abundance) in at least 2 samples (n = 1569 ASVs excluded).
#> phyloseq-class experiment-level object
#> otu_table() OTU Table: [ 224 taxa and 287 samples ]
#> sample_data() Sample Data: [ 287 samples by 5 sample variables ]
#> tax_table() Taxonomy Table: [ 224 taxa by 8 taxonomic ranks ]Use a filter described by Subramanian et al. (Nature, 2014) to filter phyloseq-objects.
Extract metadata
ps_NP %>%
meta_to_df() %>%
head()
#> sample_id seq_id visit_name age birth_mode bf_group_3m_yn
#> 1 121142010012 M001 w1 8 vag 1
#> 2 121144010012 M001 m1 30 vag 1
#> 3 121150010012 M001 y1 364 vag 1
#> 4 121142010032 M003 w1 7 vag 1
#> 5 121144010032 M003 m1 32 vag 1
#> 6 121150010032 M003 y1 361 vag 1meta_to_df() convers the sample_data()-objects within a
phyloseq-object into a tidyverse-formatted data table. Also try
otu_tab_to_df() and ps_to_df()
Prepare for plotting
ps_NP %>%
prep_bar(n = 5) %>%
head()
#> # A tibble: 6 x 8
#> sample_id seq_id visit_name age birth_mode bf_group_3m_yn OTU value
#> <fct> <chr> <ord> <dbl> <chr> <fct> <fct> <dbl>
#> 1 121142010… M001 w1 8 vag 1 *Moraxella… 7
#> 2 121142010… M001 w1 8 vag 1 *Staphyloc… 966
#> 3 121142010… M001 w1 8 vag 1 *Corynebac… 0
#> 4 121142010… M001 w1 8 vag 1 *Dolosigra… 9616
#> 5 121142010… M001 w1 8 vag 1 *Haemophil… 0
#> 6 121142010… M001 w1 8 vag 1 Residuals 19322The function prep_bar() converts your phyloseq-object into an object
that can be readily used for plotting. It includes all metadata-columns,
a column ‘OTU’ (with formatted OTU-names) and a column ‘value’ with read
counts/relative abundances (depending on the format of the otu_table).
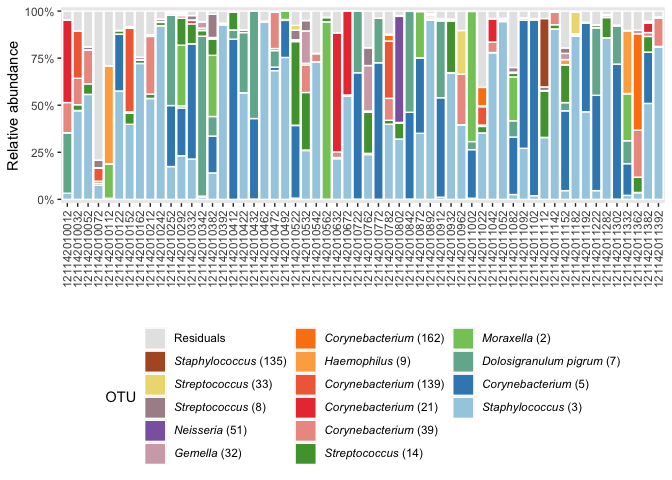
Barplot
ps_NP %>%
subset_samples(., birth_mode == "vag" & visit_name == "w1") %>% # select samples
to_RA %>%
create_bar()Note this plotting function can be used in conjunction with other
ggplot2-functions/extensions, such as
coord_flip()/ggforce::facet_col() and facet_wrap().