This repository contains code for the R package seismicGWAS a method for calculating cell type-trait associations given GWAS and single cell RNA-sequencing data.
Disentangling associations between complex traits and cell types with seismic. Lai Q, Dannenfelser R, Roussarie JP, Yao V. BioRxiv. April 2024.
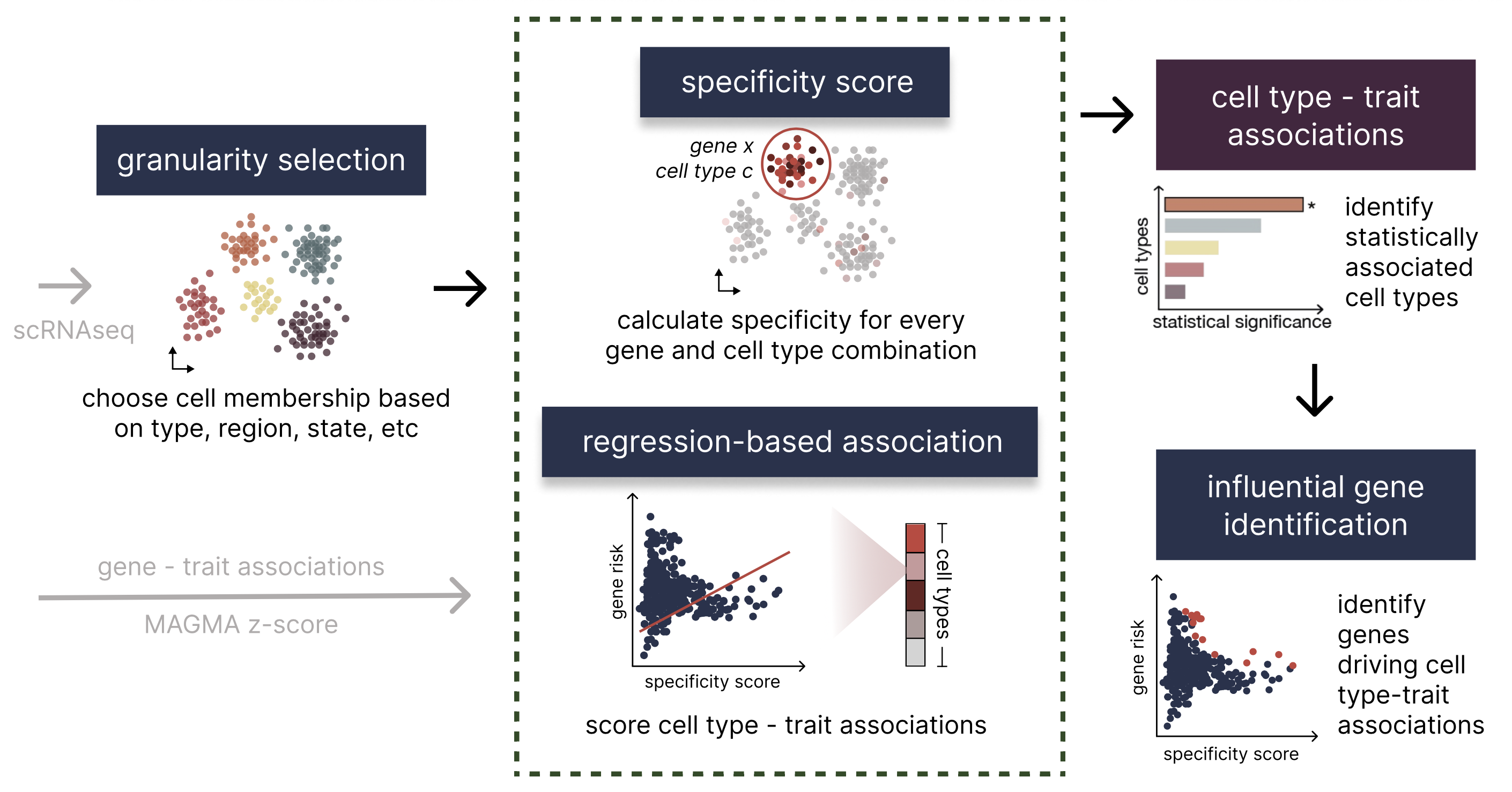
Integrating single-cell RNA sequencing (scRNA-seq) with Genome-Wide Association Studies (GWAS) can help reveal GWAS-associated cell types furthering our understanding of the cell-type-specific biological processes underlying complex traits and disease. In order to rapidly and accurately pinpoint associations, we develop a novel framework, seismic, which characterizes cell types using a new specificity score. As part of the seismic framework, the specific genes driving cell type-trait associations can easily be accessed and analyzed, enabling further biological insights. The following figure depicts a high level overview of this process.
To install the seismic package first clone the seismic repo and then use devtools within R to point to seismic and install.
devtools::install(path_to_seismic_folder)
library('seismicGWAS')Below we quickly show how to use seismicGWAS to calcuate cell
type-trait associations for the sample data included in the package.
Full usage instructions, including a walk through of all major functions
can be found in the vignette.
# calculate cell type specificity scores using included sample data
tmfacs_sscore <- calc_specificity(tmfacs_sce_small, ct_label_col='cluster_name')
# convert mouse gene identifiers to human ones that match data in GWAS summary data
# from MAGMA
tmfacs_sscore_hsa <- translate_gene_ids(tmfacs_sscore, from='mmu_symbol')
# calculate cell type-trait associations for type 2 diabetes
get_ct_trait_associations(tmfacs_sscore_hsa, t2d_magma)
# find the influential genes for a significant cell type-trait association
# in type 2 diabetes
find_inf_genes("Pancreas.beta cell", tmfacs_sscore_hsa, t2d_magma)