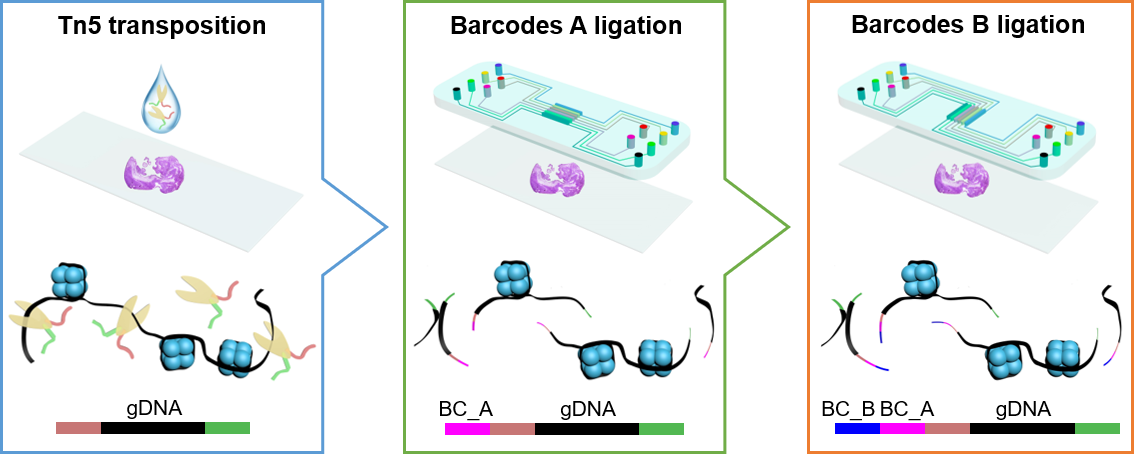
Spatial-ATAC-seq: spatially resolved chromatin accessibility profiling of tissue at genome scale and cellular level
Next Generation Sequencing (NGS) was performed using the Illumina HiSeq 4000 sequencer (pair-end 150 bp mode).
Read 1: contains the spatial Barcode A and Barcode B
Read 2: contains the genome sequences
Raw read 1 -> New Read 1 + New Read 2
-
New Read 1: contains the genome sequences
-
New Read 2: contains the spatial Barcode A and Barcode B
Raw read 2 -> New Read 3
Reformatting raw data was implemented by BC_process.py in the Data_preprocessing folder.
The reformated data was processed using Cell Ranger ATAC v1.2 with following references:
Mouse reference (mm10):
curl -O https://cf.10xgenomics.com/supp/cell-atac/refdata-cellranger-atac-mm10-1.2.0.tar.gz
Human reference (GRCh38):
curl -O https://cf.10xgenomics.com/supp/cell-atac/refdata-cellranger-atac-GRCh38-1.2.0.tar.gz
A preprocessing pipeline we developed using Snakemake workflow management system is in the Data_preprocessing folder. To run the pipeline, use the command:
sbatch Snakemake.sh
The data visualization were implemented with ArchR v1.0.1 and Seurat v3.2 package (Data_visualization folder).
Brief descriptions of analysis scripts:
metadata_files_for_Seurat_spatial.ipynb: Generate metadata files that were compatible with Seurat workflow for spatial datasets.
archR.R: Data normalization and dimensionality reduction, identifying the marker genes, peak calling, deviatons enrichment anaylsis, bulk sample projection, and pseudotime analysis.
spatial_data_visualization.R: Visualize spatially resolved data on tissue sections.
GO_enrichment_analysis.R: GO enrichment analysis for marker genes.
integrative_data_analysis.R: Integrative data analysis with scRNA-seq reference datasets.