Improving Self-Supervised Learning with Hardness-aware Dynamic Curriculum Learning: An Application to Digital Pathology
by Chetan L. Srinidhi and Anne L. Martel
-
Official repository for Improving Self-supervised Learning with Hardness-aware Dynamic Curriculum Learning: An Application to Digital Pathology. Accepted in ICCV, 2021, CDpath workshop, October, 2021. [Conference proceedings] [arXiv preprint]
In this work, we attempt to improve self-supervised pretrained representations through the lens of curriculum learning by proposing a hardness-aware dynamic curriculum learning (HaDCL) approach. To improve the robustness and generalizability of SSL, we dynamically leverage progressive harder examples via easy-to-hard and hard-to-very-hard samples during mini-batch downstream fine-tuning. We discover that by progressive stage-wise curriculum learning, the pretrained representations are significantly enhanced and adaptable to both in-domain and out-of-domain distribution data.
We carry out extensive validation experiments on three histopathology benchmark datasets on both patch-wise and slide-level classification tasks:
- Camelyon 16
- Memorial Sloan Kettering Cancer Center (MSKCC), Breast Metastases dataset
- Colorectal Polyps classification
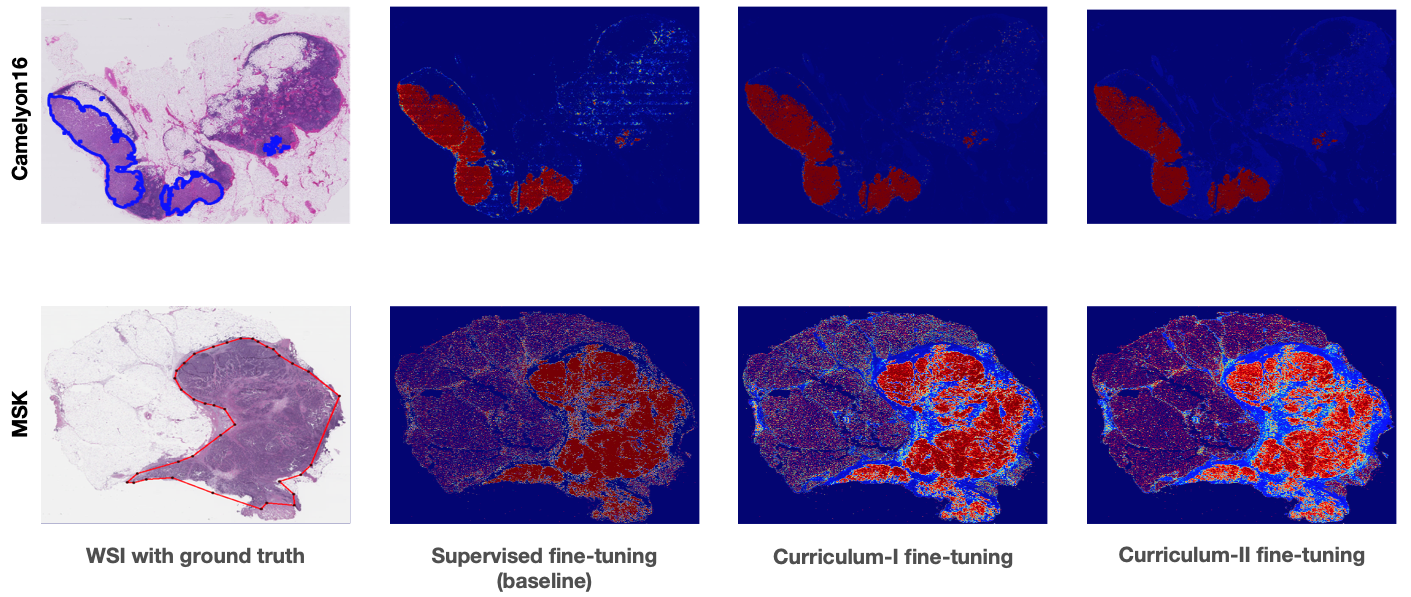
- Predicted tumor probability heat-maps on Camelyon16 (in-domain) and MSKCC (out-of-domain) test sets
Core implementation:
- Python 3.7+
- Pytorch 1.7+
- Openslide-python 1.1+
- Albumentations 1.8+
- Scikit-image 0.15+
- Scikit-learn 0.22+
- Matplotlib 3.2+
- Scipy, Numpy (any version)
- Camelyon16: to download the dataset, check this link :
https://camelyon16.grand-challenge.org - MSKCC: to download the dataset, check this link :
https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=52763339 - MHIST: to download the dataset, check this link :
https://bmirds.github.io/MHIST/
The model training consists of three steps:
- Self-supervised pretraining (i.e., Our earlier proposed method
Resolution sequence prediction (RSP)and Momentum Contrast (MoCo)) - Curriculum-I fine-tuning (
easy-to-hard) - Curriculum-II fine-tuning (
hard-to-very-hard)
In this work, we build our approach on our previous work "Self-Supervised driven Consistency Training for Annotation Efficient Histopathology Image Analysis. Please, refer to our previous repository for pretraining details on whole-slide-images. We have included the pretrained model (cam_SSL_pretrained_model.pt) for Camelyon16, found in the "models" folder.
2, 3. Fine-tuing of pretrained models on the target task using hardness-aware dynamic curriculum learning
- Download the self-supervised pretrained model from the models folder.
- Download the desired dataset; you can simply add any other dataset that you wish.
- For slide-level classification tasks, run the following command by the desired parameters. The arguments can be set in the corresponding files.
python ft_Cam_SSL.py // Supervised fine-tuning on Camelyon16
python ft_Cam_SSL_CL_I.py // Curriculum-I fine-tuning on Camelyon16
python ft_Cam_SSL_CL_II.py // Curriculum-II fine-tuning on Camelyon16We have included the fine-tuned models (cam_curriculum_I_finetuned_model.pt, cam_curriculum_II_finetuned_model.pt) for Camelyon16, found in the "models" folder. These models can be used to test the predictions on Camelyon16 and MSKCC test sets.
- For patch-level classification tasks, run the following command by the desired parameters. The arguments can be set in the corresponding files.
python ft_MHSIT_SSL.py // Supervised fine-tuning on MHIST
python ft_MHSIT_SSL_CL_I.py // Curriculum-I fine-tuning on MHIST
python ft_MHSIT_SSL_CL_II.py // Curriculum-II fine-tuning on MHISTThe test performance is validated at two stages:
- Patch-level predictions: The patch-level predictions can be performed to generate tumor probability heat-maps on Camelon16 and MSKCC datasets. Note: MSKCC doesn't contain any training images and hence, we use this dataset as an external validation set to test our method's performance on out-of-distribution data.
prob_map_generation.py // tumor probability heat-map on Camelyon16, MSKCC
eval_MHIST.py // patch-wise predictions on MHIST - Random-forest-based slide-level classifier: The final slide-level predictions can be performed using scripts inside the "Slide_Level_Analysis" folder.
extract_feature_heatmap.py // to extract geometrical features from the heatmap predictions of the previous stage
wsi_classification.py // Random-forest based slide-level classifierOur code is released under MIT license.
If you find our work useful in your research or if you use parts of this code please consider citing our papers:
@article{srinidhi2021improving,
title={Improving Self-supervised Learning with Hardness-aware Dynamic Curriculum Learning: An Application to Digital Pathology},
author={Srinidhi, Chetan L and Martel, Anne L},
journal={arXiv preprint arXiv:2108.07183},
year={2021}
}
@article{srinidhi2021self,
title={Self-supervised driven consistency training for annotation efficient histopathology image analysis},
author={Srinidhi, Chetan L and Kim, Seung Wook and Chen, Fu-Der and Martel, Anne L},
journal={arXiv preprint arXiv:2102.03897},
year={2021}
}
This work was funded by Canadian Cancer Society and Canadian Institutes of Health Research (CIHR). It was also enabled in part by support provided by Compute Canada (www.computecanada.ca).
Please direct any questions or comments to me; I am happy to help in any way I can. You can email me directly at chetan.srinidhi@utoronto.ca.