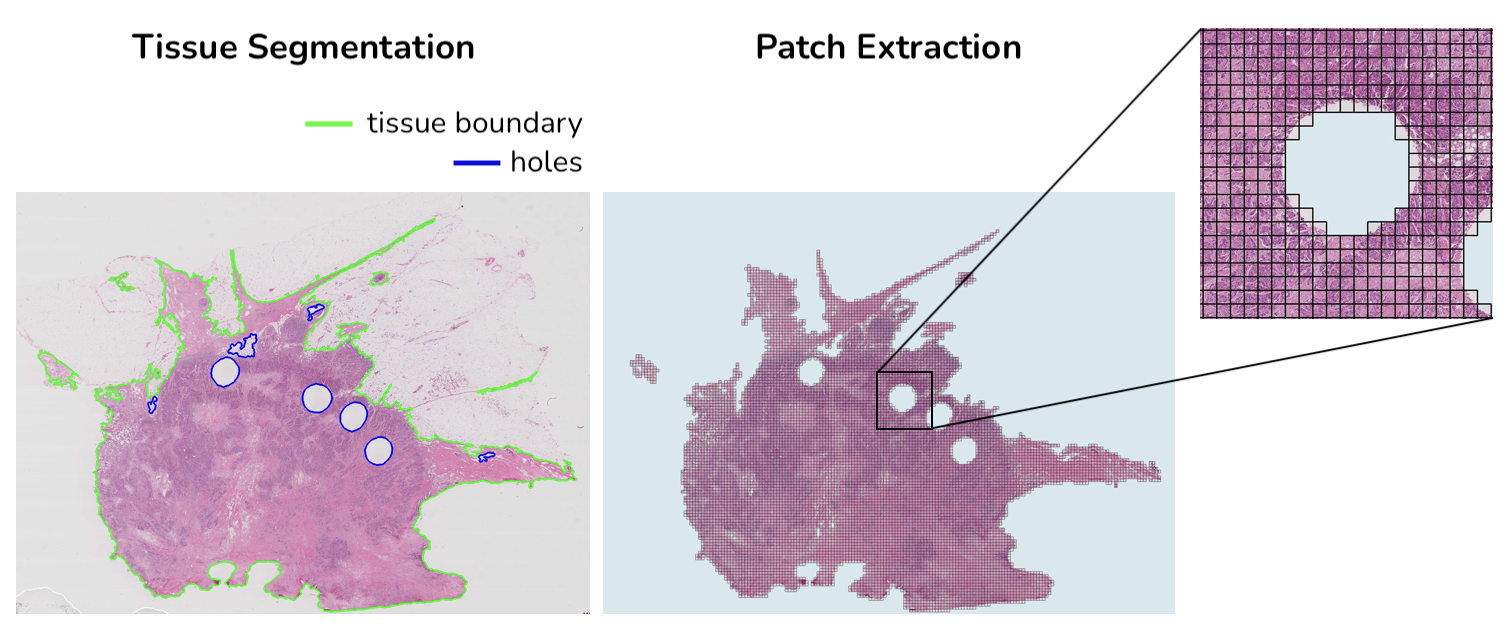
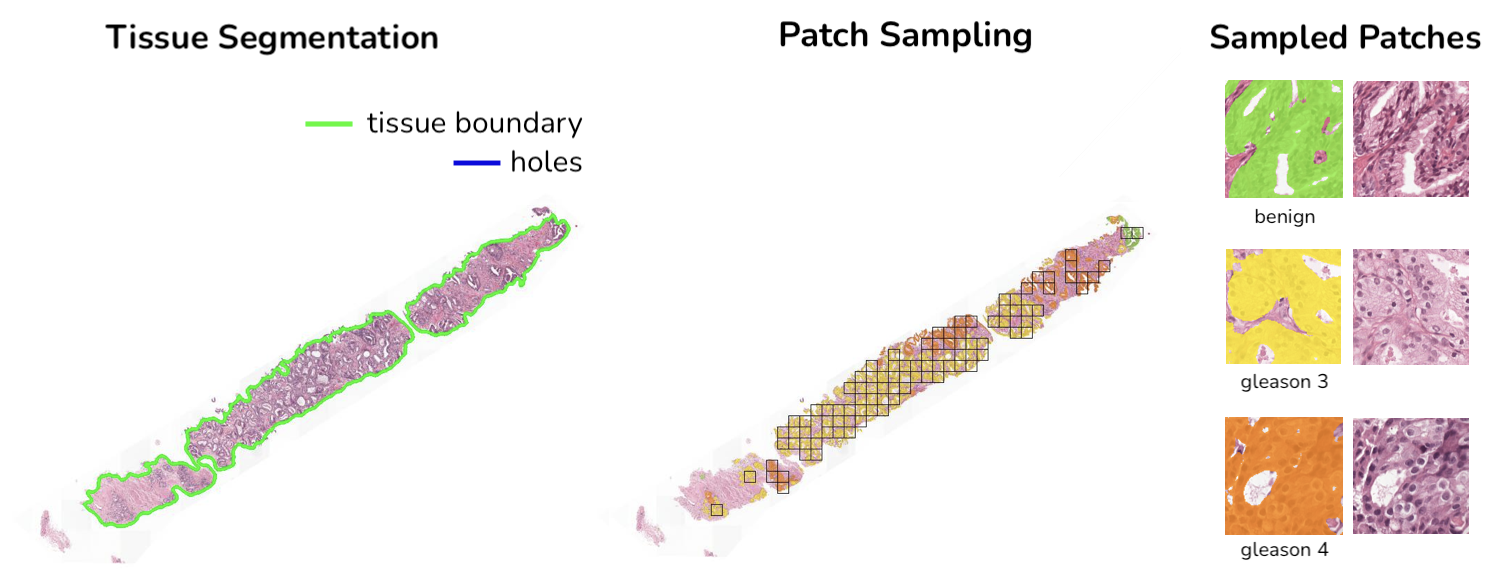
HS2P is an open-source project largely based on CLAM tissue segmentation and patching code.
install requirements via pip3 install -r requirements.txt
- [Optional] Configure wandb
If you want to benefit from wandb logging, you need to follow these simple steps:
- grab your wandb API key under your profile and export
- run the following command in your terminal:
export WANDB_API_KEY=<your_personal_key> - change wandb paramters in the configuration file under
config/(setenabletoTrue)
- Create a .csv file containing paths to the desired slides:
slide_id,slide_path
slide_id_1,path/to/slide_1.tif
slide_id_2,path/to/slide_2.tif
...
You can optionally provide paths to pre-computed segmentation masks under the 'segmentation_mask_path' column
slide_id,slide_path,segmentation_mask_path
slide_id_1,path/to/slide_1.tif,path/to/slide_1_mask.tif
slide_id_2,path/to/slide_2.tif,path/to/slide_2_mask.tif
...
- Create a configuration file under
config/extraction/
A good starting point is to use the default configuration file config/extraction/default.yaml where parameters are documented.
- Run the following command to kick off the algorithm:
python3 patch_extraction.py --config-name <config_filename>
- Depending on which flags have been set to True, it will produce (part of) the following results:
Patch extraction output
hs2p/
├── output/<experiment_name>/
│ ├── masks/
│ │ ├── slide_id_1.jpg
│ │ ├── slide_id_2.jpg
│ │ └── ...
│ ├── patches/<patch_size>/<format>/
│ │ ├── slide_id_1/
│ │ │ ├── slide_id_1.h5
│ │ │ └── imgs/
│ │ │ ├── x0_y0.<format>
│ │ │ ├── x1_y0.<format>
│ │ │ └── ...
│ │ ├── slide_id_2/
│ │ └── ...
│ ├── visualization/
│ │ └── <patch_size>/
│ │ ├── slide_id_1.jpg
│ │ ├── slide_id_2.jpg
│ │ └── ...
│ ├── tiles.csv
│ └── process_list.csv
masks/ will contain a downsampled view of the slide with tissue segmentation overlayed
visualization/ will contain a downsampled view of the slide where extracted patches are highlighted
tiles.csv contain patching information for each slide that ended up having patches extracted:
slide_id,tile_size,spacing,level,level_dim,x,y,contour
slide_id_1,2048,0.5,0,"(10496, 20992)",752,5840,0
...
Extracted patches will be saved as x_y.jpg where x and y represent the true location in the slide at level 0:
- if spacing at level 0 is
0.25and you extract [256, 256] patches at spacing0.25, two consecutive patches will be distant from256pixels (either alongxoryaxis) - if spacing at level 0 is
0.25and you extract [256, 256] patches at spacing0.5, two consecutive patches will be distant from512pixels (either alongxoryaxis)
- [Optional] Configure wandb
see above
- Create a .csv file containing paths to the desired slides & associated annotation masks:
slide_id,slide_path,annotation_mask_path
slide_id_1,path/to/slide_1.tif,path/to/slide_1_annot_mask.tif
slide_id_2,path/to/slide_2.tif,path/to/slide_2_annot_mask.tif
...
In the same way as for patch extraction, you can optionally provide paths to pre-computed segmentation masks under the 'segmentation_mask_path' column.
- Create a configuration file under
config/sampling/
A good starting point is to use the default configuration file config/sampling/default.yaml where parameters are documented.
- Run the following command to kick off the algorithm:
python3 patch_sampling.py --config-name <config_filename>
- Depending on your config, it will produce (part of) the following results:
Patch sampling output
hs2p/
├── output/<experiment_name>/
│ ├── annotation_mask/
│ │ ├── slide_id_1.jpg
│ │ ├── slide_id_2.jpg
│ │ └── ...
│ ├── segmentation_mask/
│ │ ├── slide_id_1.jpg
│ │ ├── slide_id_2.jpg
│ │ └── ...
│ ├── patches/
│ │ ├── raw/
│ │ │ ├── category_1/
│ │ │ │ ├── slide_id_1_x0_y0.<format>
│ │ │ │ ├── slide_id_1_x1_y0.<format>
│ │ │ │ └── ...
│ │ │ ├── category_2/
│ │ │ └── ...
│ │ ├── mask/
│ │ │ ├── category_1/
│ │ │ │ ├── slide_id_1_x0_y0_mask.<format>
│ │ │ │ ├── slide_id_1_x1_y0_mask.<format>
│ │ │ │ └── ...
│ │ │ ├── category_2/
│ │ │ └── ...
│ │ └── h5/
│ │ ├── slide_id_1.h5
│ │ ├── slide_id_2.h5
│ │ └── ...
│ ├── visualization/
│ │ ├── slide_id_1.jpg
│ │ ├── slide_id_2.jpg
│ │ └── ...
│ └── sampled_tiles.csv
annotation_mask/ will contain a downsampled view of the slide with corresponding annotation mask overlayed
segmentation_mask/ will contain a downsampled view of the slide with tissue segmentation overlayed
visualization/ will contain a downsampled view of the slide where sampled patches are highlighted
sampled_patches.csv contain information for each patch that ended up being extracted:
slide_id,category,x,y,pct
slide_id_1,category_1,3488,2512,0.8203125
...
Again, extracted patches will be saved as x_y.jpg where x and y represent the true location in the slide at level 0.
If, for some reason, the experiment crashes, you should be able to resume from last processed slide simply by turning the resume parameter in your config file to True, keeping all other parameters unchanged.
If the generated visualization are noisy, you'll need to change libpixman version. Running the following command should fix this issue:
wget https://www.cairographics.org/releases/pixman-0.40.0.tar.gz
tar -xf pixman-0.40.0.tar.gz
cd pixman-0.40.0
./configure
make
make install
export LD_PRELOAD=/usr/local/lib/libpixman-1.so.0.40.0