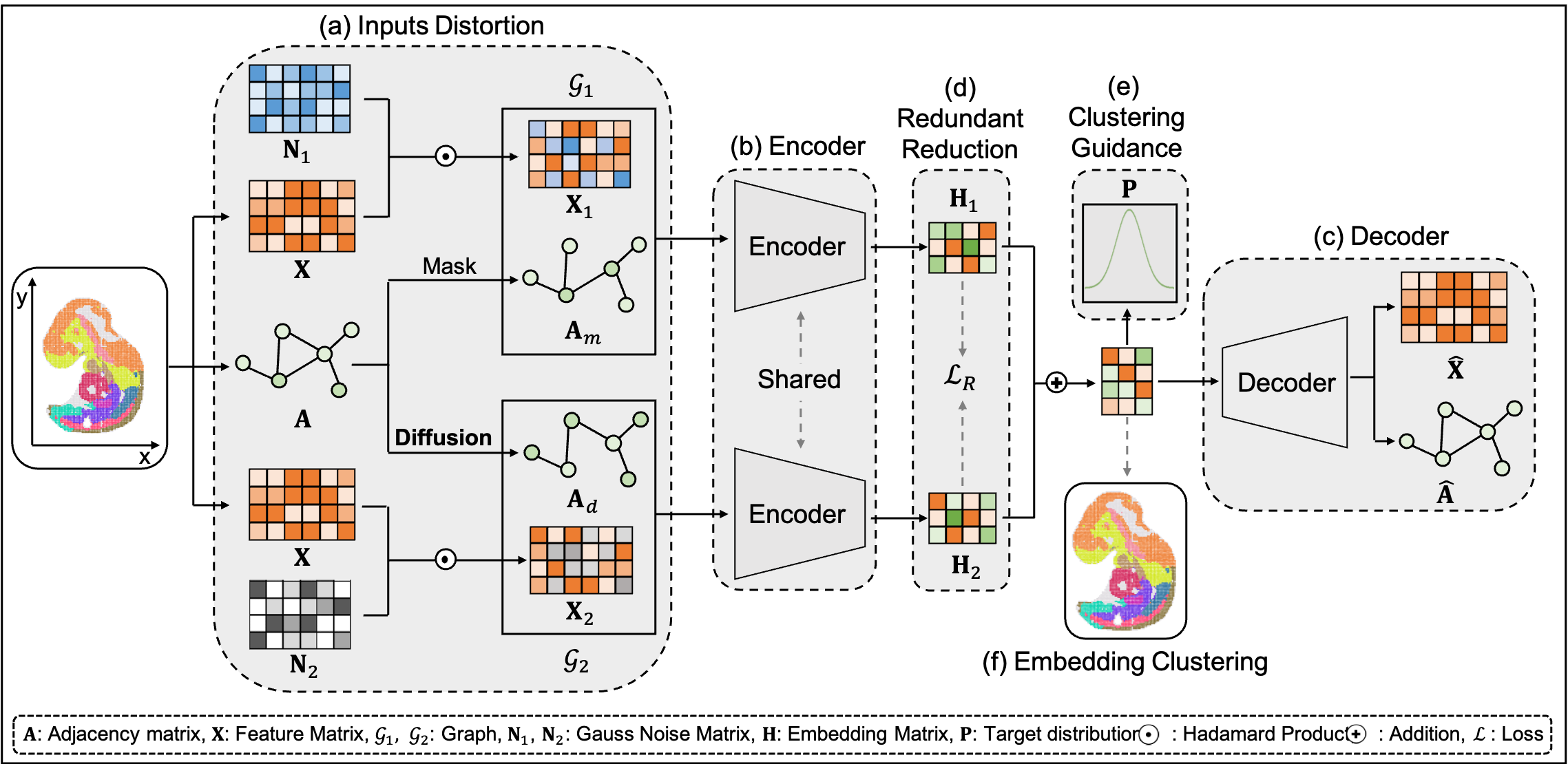
SGAE: Deciphering spatial domains from spatially resolved transcriptomics with Siamese Graph Autoencoder
Spatial transcriptomics (ST) is a newly emerging field that facilitates a comprehensive characterization of tissue organization and architecture. By profiling the spatially-resolved gene expression patterns, ST technologies allow scientists to gain an in-depth understanding of the complex cellular dynamics and within tissue. Graph neural network (GNN) based methods usually suffer from representations collapse, which tends to map spatial spots into same representation. To address this issue, we proposed a Siamese Graph Autoencoder (SGAE) framework to learn discriminative spot representation and decipher accurate spatial domains. SGAE outperformed those spatial clustering methods across multiple platforms derived datasets based on the evaluation of ARI, FMI, NMI. Moreover, the clustering results derived from the SGAE model can be further utilized in 3D Drosophila Embryo reconstruction.
### Python enviroment constructed by Conda
conda create -n SGAE python=3.8
conda activate SGAE
pip install -r requirements.txtWe also upload our code to Code Ocean. Please check it for easier compilation.
cd SGAE
python3 run_sgae.py --n_epochs 1000 --name xxx --data_file xxx.h5adPlease specify your own data name and data_file via the arguments showed above. You can also check the tutorial below to get a quick start.
We used data from various of platform and samples to benchmark our method. Here is a table for the data mentioned in article.
| Dataset | Platform | Samples | Species | Tissue | Source |
|---|---|---|---|---|---|
| DLPFC | 12 | 10X Visium | Human | Dorsolateral prefrontal cortex | http://research.libd.org/spatialLIBD |
| MG | 1 | seqFISH | Mouse | Gastrulation | https://crukci.shinyapps.io/SpatialMouseAtlas/ |
| MC | 1 | MERFISH | Mouse | Cortex data | https://doi.brainimagelibrary.org/ |
| MOB | 1 | SLIDE-seq v2 | Mouse | Olfactory bulb | https://singlecell.broadinstitute.org/single_cell/study/SCP815/highly-sensitive-spatial-transcriptomics-at-near-cellular-resolution-with-slide-seqv2#study-summary |
| DE | 16 | Stereo-seq | Drosophila | Embryo | https://db.cngb.org/stomics/flysta3d/spatial/ |
| MB | 1 | Stereo-seq | Mouse | Brain | https://zenodo.org/record/7340795 |
The foundation functions of SGAE is deposited at models directory.
-
General setting
Parameter Type Defination Default name str name to save result to indicate the data or sample dblp modelname str name to save result to indicate the model SGAE project_dir str directory to save result ./ cuda bool whether to use GPU True gpu_id str choose a specific GPU 0 seed int determine random seed 1 n_clusters int number of clustering 20 -
Graph setting
Parameter Type Defination Default k_nn int number of neighbors to construct graph 3 alpha_value float alpha value for graph diffusion 0.2 -
Training setting
Parameter Type Defination Default n_epochs int total epoch 1000 patience float denote the early stopping point 0.2 batch_size int the size of a single batch 256 lr float learning rate 1e-4 lambda_value float weight for clustering guidance loss 10
Reproduce the result of article via run_case.py.
-
DLPFC:
python3 run_case.py --n_epochs 1000 --dataset dlpfc
-
seqFISH Mouse Gastrulation:
python3 run_case.py --n_epochs 1000 --dataset seqfish
-
MERFISH Mouse Cortex data:
python3 run_case.py --n_epochs 1000 --dataset merfish
-
SLIDE-seq v2 Mouse Olfactory bulb:
python3 run_case.py --n_epochs 1000 --dataset slideseq
-
Stereo-seq Drosophila Embryo:
python3 run_case.py --n_epochs 1000 --dataset drosophila_14_16 python3 run_case.py --n_epochs 1000 --dataset drosophila_16_18 python3 run_case.py --n_epochs 1000 --dataset drosophila_l1
-
Stereo-seq Mouse Brain:
python3 run_case.py --n_epochs 1000 --dataset mousebrain
Any questions or suggestions on EAGS are welcomed! Please report it on issues, or contact Lei Cao (caolei2@genomics.cn) or Shuangsang Fang (fangshuangsang@genomics.cn). We recommend using STOmics Cloud Platform to access and use it.