Cross-slice Spatial Transcriptome Domain Analysis.
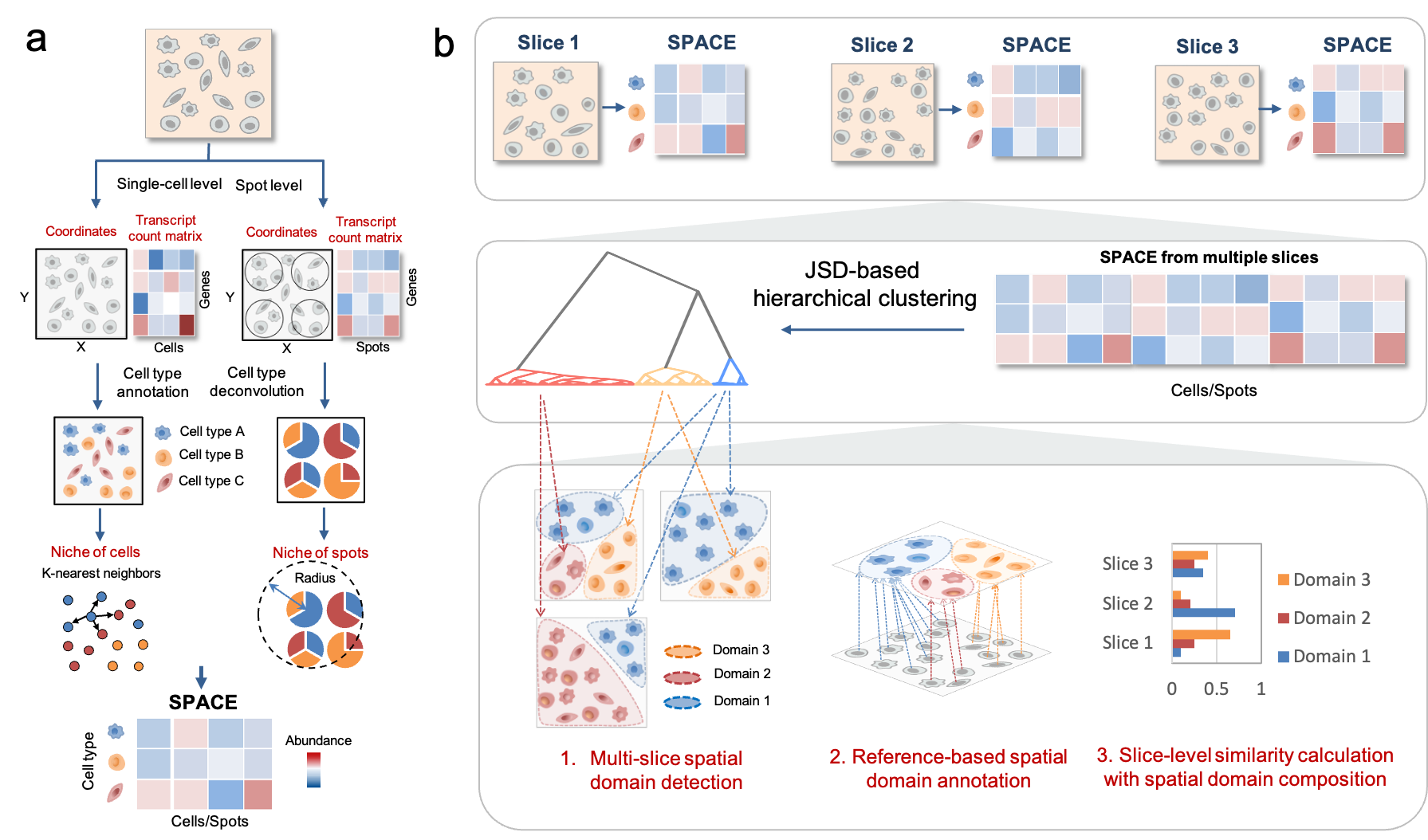 Fig 1. Workflow of SpaDo. a. Calculating the SPACE for both single-cell resolution and spot resolution spatial transcriptomic data. SPACE: SPatially Adjacent Cell type Embedding. b. Three functions involved in cross-slice spatial domain analysis: multi-slice domain detection, reference-based spatial domain annotation, and cross-slice clustering analysis by consideration of spatial domain composition. JSD, Jensen Shannon Divergence.
Fig 1. Workflow of SpaDo. a. Calculating the SPACE for both single-cell resolution and spot resolution spatial transcriptomic data. SPACE: SPatially Adjacent Cell type Embedding. b. Three functions involved in cross-slice spatial domain analysis: multi-slice domain detection, reference-based spatial domain annotation, and cross-slice clustering analysis by consideration of spatial domain composition. JSD, Jensen Shannon Divergence.
With the rapid advancements in spatial transcriptome sequencing, multiple tissue slices are now available, enabling the integration and interpretation of spatial cellular landscapes. Herein, we introduce SpaDo, a tool for cross-slice spatial domain detection, annotation, and downstream analysis at both single-cell and spot resolutions. SpaDo includes modules for cross-slice spatial domain detection, reference-based annotation, and cross-slice clustering. We demonstrated SpaDo's effectiveness with over 40 multi-slice spatial transcriptome datasets from 7 sequencing platforms. Our findings highlight SpaDo's potential to reveal novel biological insights in multi-slice spatial transcriptomes.
-
SpaDo package can be installed from Github using devtools packages with R>=4.0.5.
library(devtools) install_github("bm2-lab/SpaDo")
See Tutorials and Demo datasets
Bin Duan, Shaoqi Chen, Xiaojie Cheng, Qi Liu. Cross-slice Spatial Transcriptome Domain analysis with SpaDo.