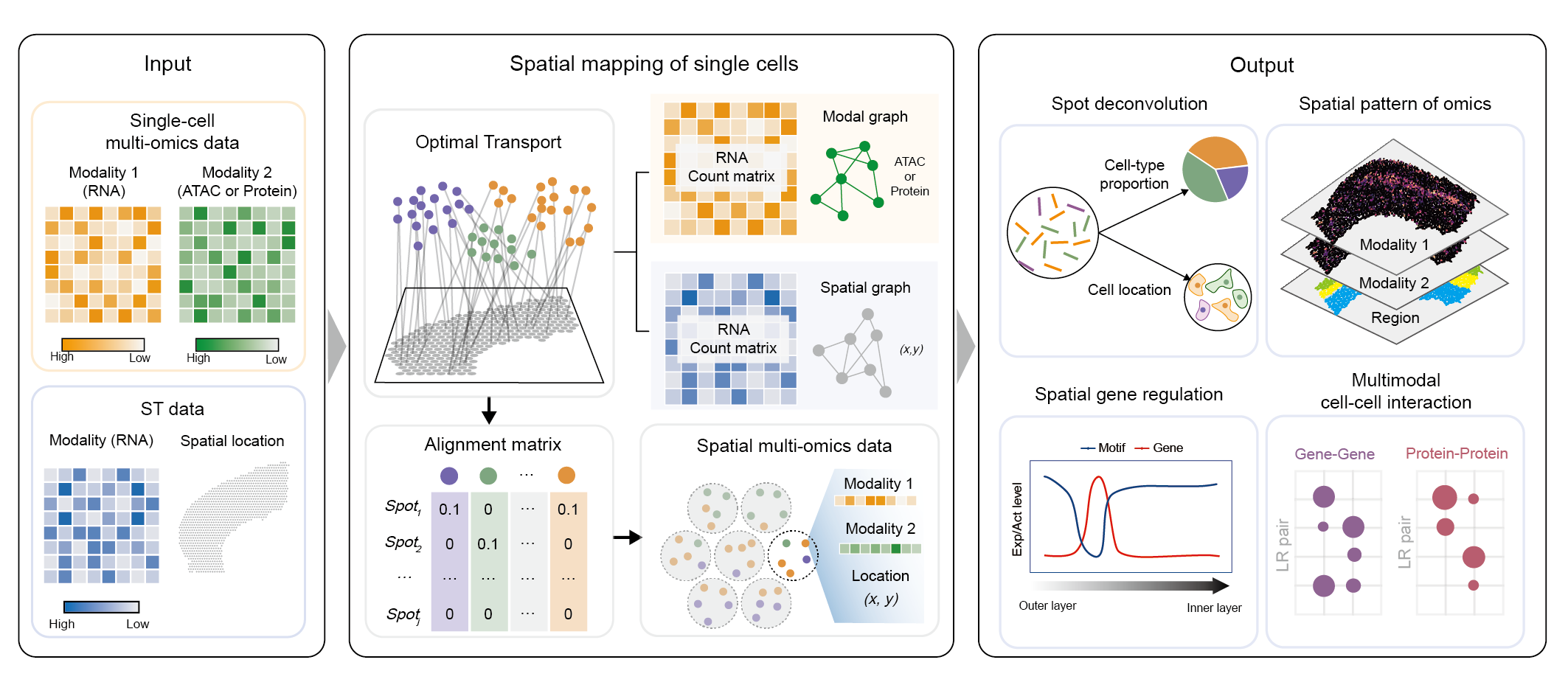
SpaTrio is a computational tool based on optimal transport that can align single-cell multi-omics data in space while preserving the spatial topology of the tissue section and local geometry of modality
This toolkit is written in both R and Python programming languages. The core optimal transport algorithm is implemented in Python, while the initial data preparation and downstream multimodal analysis are written in R.
# We recommend using Anaconda, and then you can create a new environment.
# Create and activate Python environment
conda create -n spatrio python=3.8
conda activate spatrio
# Install requirements
cd SpaTrio-main
pip install -r requirements.txt
# Install spatrio
python setup.py build
python setup.py install
install.packages("doParallel")
BiocManager::install("ConsensusClusterPlus")
# Install SpaTrio package from local file
install.packages("SpaTrio_1.0.0.tar.gz", repos = NULL, type = "source")
To use SpaTrio we require formatted .csv files as input (i.e. read in by pandas).
- multi_rna.csv/spatial_rna.csv (The gene expression matrix of cells/spots)
| Cell1 | ··· | Celln | |
|---|---|---|---|
| Gene1 | 0 | ··· | 1 |
| ··· | ··· | ··· | ··· |
| Genem | 2 | ··· | 1 |
- multi_meta.csv/spatial_meta.csv (The meta information matrix of cells/spots)
| id | type | |
|---|---|---|
| Cell1 | Cell1 | A |
| ··· | ··· | ··· |
| Celln | Celln | B |
- emb.csv (The low-dimensional embedding matrix of cells)
| emb1 | ··· | embk | |
|---|---|---|---|
| Cell1 | 1.997 | ··· | -0.307 |
| ··· | ··· | ··· | ··· |
| Celln | 2.307 | ··· | 2.119 |
- pos.csv (The spatial location matrix of spots)
| x | y | |
|---|---|---|
| Cell1 | 0.28 | 10.65 |
| ··· | ··· | ··· |
| Celln | 5.98 | 2.16 |
At the same time, we also support additional specifications of the number of cells in each spot.
- expected_num.csv (The number of cells contained in each spot)
| cell_num | |
|---|---|
| Spot1 | 5 |
| ··· | ··· |
| Spotj | 2 |
In some examples of simulated data, the number of cell types in the spot is given (ref_counts.csv). These data will be converted to expected_num for use.
- ref_counts.csv (The number of celltypes contained in each spot)
| Celltype 1 | ··· | Celltype i | |
|---|---|---|---|
| Spot1 | 0 | ··· | 2 |
| ··· | ··· | ··· | ··· |
| Spotj | 1 | ··· | 0 |
We have included two test datasets (demo1 & demo2) in the tutorial/data/ of this repository as examples to show how to use SpaTrio to align cells to space.
Simulated data in the stripe pattern:
Simulated data in the ring pattern:
More importantly, we support directly calling the core functions written in Python from the R language to facilitate downstream analysis.
DBiT-seq mouse embryo datasets (Google Drive):
10x Visium+ADT mouse liver datasets (Google Drive):
We have applied SpaTrio on different tissues of multiple species, here we give step-by-step tutorials for all application scenarios. And preprocessed datasets used can be downloaded from Google Drive.
-
Using SpaTrio to reconstruct and analyze single-cell multi-modal data of mouse cerebral cortex
-
Using SpaTrio to reconstruct and analyze single-cell multi-modal data of human steatosis liver
-
Using SpaTrio to reconstruct and analyze single-cell multi-modal data of human breast cancer
Should you have any questions, please feel free to contact the author of the manuscript, Mr. Penghui Yang (yangph@zju.edu.cn).
Penghui Yang, et al. Revealing spatial multimodal heterogeneity in tissues with SpaTrio, Cell Genomics, 2023, https://doi.org/10.1016/j.xgen.2023.100446.