A simple plugin to visualize multi-plexed FISH data to explore the 3D genome organization and make interactive exploration with multi-modal data hosted in Nucleome Browser.
This napari plugin was generated with Cookiecutter using @napari's cookiecutter-napari-plugin template.
You can install napari-nucleome via pip:
pip install napari-nucleome
Napari-nucleome is a plugin for napari to visulize multi-plexed FISH data (eg., DNA seqFISH+ data generated in Takei et al., Nature 2021) in napari. This plugin allows users to perform interactive exploration of multi-modal datasets, including imaging, multi-omics and 3D genome structure models for studying the cell nucleus.
The following animation demostrates the use case of explorting DNA seqFISH+ data in napari. Users can query probes by their genomic position, switch different targets, and choose different visualization parameters.
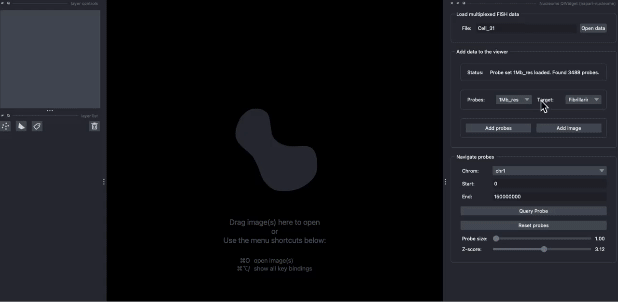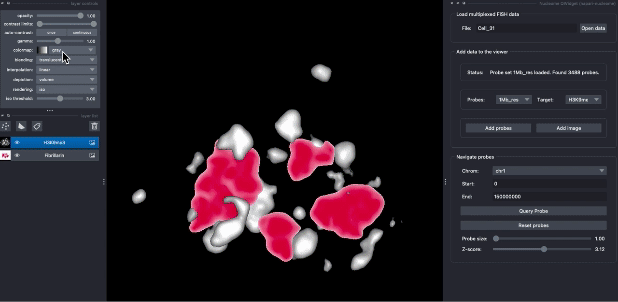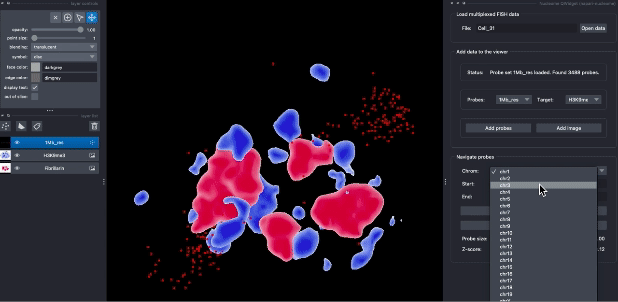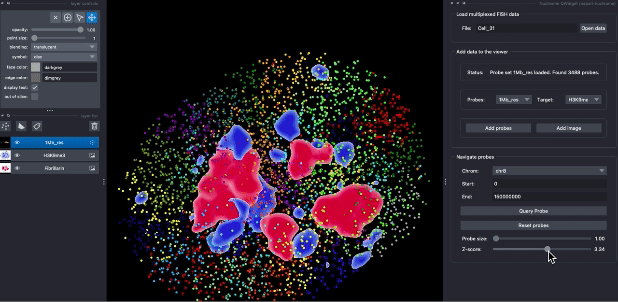
Contributions are very welcome. Tests can be run with tox, please ensure the coverage at least stays the same before you submit a pull request.
Distributed under the terms of the MIT license, "napari-nucleome" is free and open source software
If you encounter any problems, please [file an issue] along with a detailed description.



