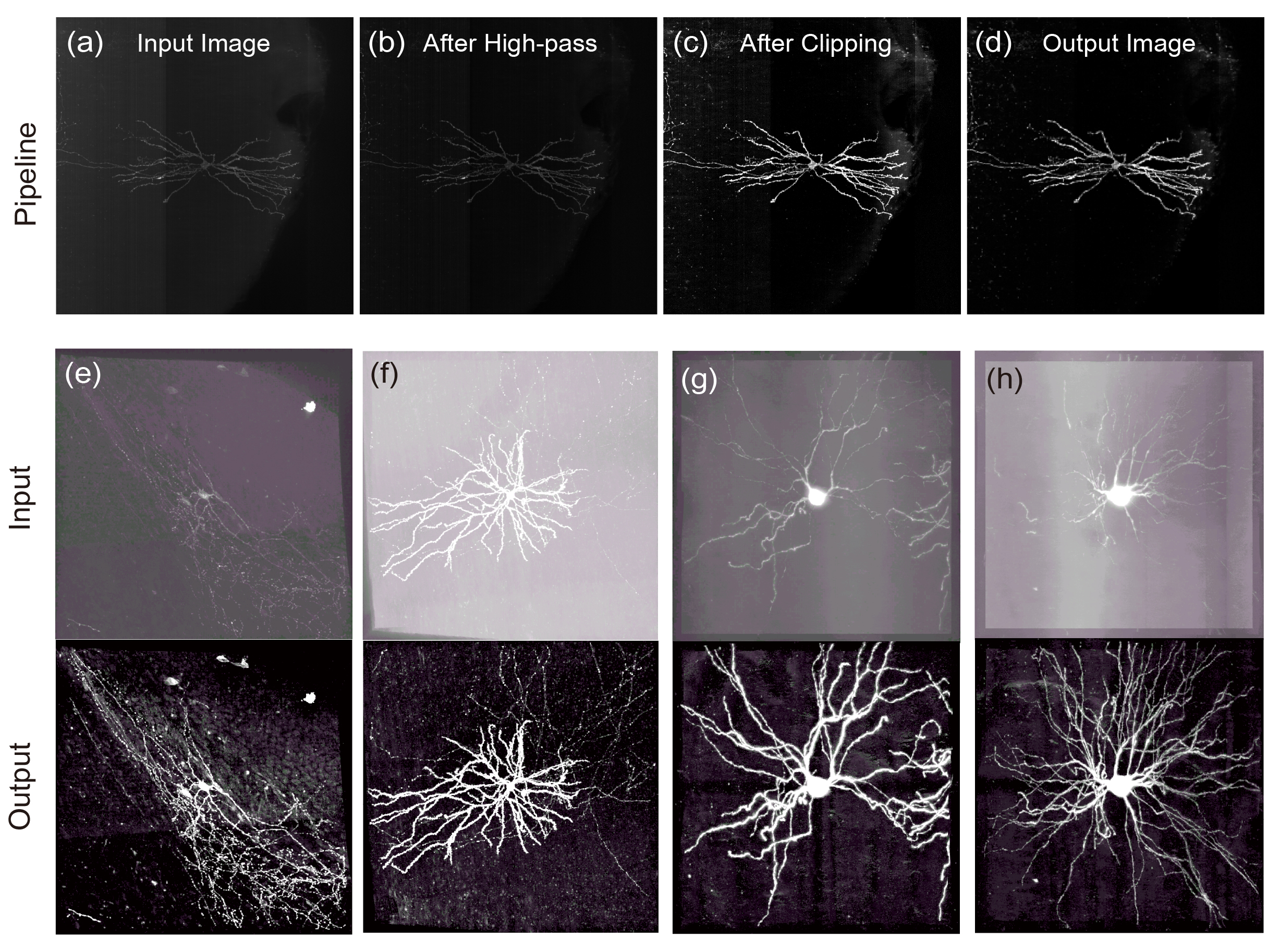
NIEND is a fast and powerful neuronal image denoising and enhancement algorithm.
from niend import standard_niend, simple_niend
from v3dpy.loaders import PBD, Raw
from utils.file_io import load_image, save_image # in this repo
img = PBD().load('xxx.v3dpbd')[0] # loaded as 4D
img = Raw().load('xxx.v3draw')[0] # loaded as 4D
img = load_image('xxx.tiff')
# standard niend: for soma position already known
# you can specify where the soma is, this works when
# the whole neuron's signal is very weak
img = standard_niend(img, # 3D numpy array indexed by z, y, x
sigma=12, pct=5,
soma=(.5, .5, .5), # center of the block
win=(32, 32, 32),
wavelet_levels=1)
# simple niend: for soma position unknown
# the result of this should be the same with `standard_niend`
# in most cases, as the neurite signal usually spans a large range
img = simple_niend(img, sigma=12, pct=5, wavelet_levels=1)
img = PBD().save('xxx.v3dpbd', img.reshape(1, *img.shape)) # saved as 4D
img = Raw().save('xxx.v3draw', img.reshape(1, *img.shape)) # saved as 4D
img = load_image('xxx.tiff')niend.py: main filtering functions, you can import the functions within for your own use.experiments: scripts performing all experiments.plot: scripts plotting figures for our paper.utils: for operating swc, computing metrics, etc.examples: image and tracing files for the same neuronal block.
More readme can be found under each folder.
Zuo-Han Zhao, Braintell, Southeast University
Yufeng Liu, Braintell, Southeast University
This paper is currently under review by Bioinformatics. Preprint is available at https://www.biorxiv.org/content/10.1101/2023.10.21.563265v1.