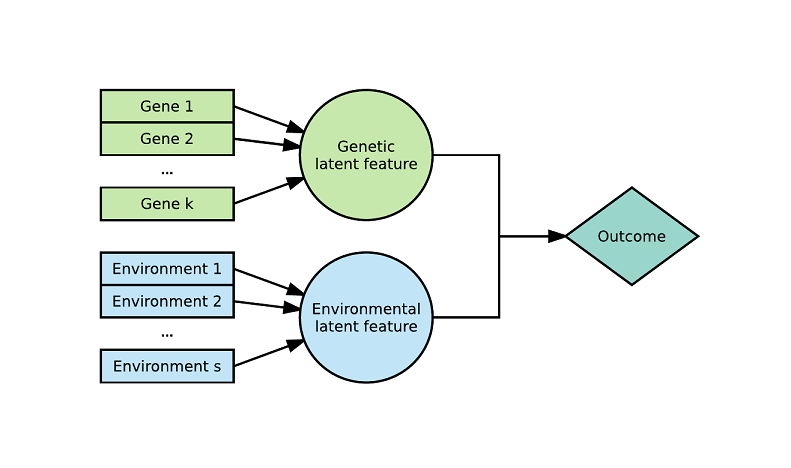
This package contains the official R implementation of the Latent Environmental & Genetic InTeraction (LEGIT) model and additional tools for gene-by-environment (GxE) testing.
If you use this software, please cite:
Jolicoeur-Martineau, A., Wazana, A., Szekely, E., Steiner, M., Fleming, A. S., Kennedy, J. L., ... & Greenwood, C. M. (2018). Alternating optimization for G× E modelling with weighted genetic and environmental scores: Examples from the MAVAN study. Psychological methods.
Paper: https://www.dropbox.com/s/swkwcnc4b4bvx8f/Jolicoeur-Martineau%20et%20al.%2C%202019_alt.pdf?dl=0
If you use the function for GxE interaction testing, please also cite:
Jolicoeur-Martineau, A., Belsky, J., Szekely, E., Widaman, K., Pluess, M., Greenwood, C., & Wazana, A. (2020). Distinguishing differential susceptibility, diathesis-stress, and vantage sensitivity: Beyond the single gene and environment model. Development and Psychopathology, 32(1), 73-83. doi:10.1017/S0954579418001438
Paper: https://www.dropbox.com/s/py5r3ye5cyl3prw/Jolicoeur-Martineau%20et%20al.%2C%202019.pdf?dl=0
The LEGIT model is an interaction model with two latent variables: a weighted sum of genetic variants (genetic score) and a weighted sum of environmental variables (environmental score). Alternating optimization is used to estimate the model parameters (https://arxiv.org/abs/1703.08111). This approach has greatly enhanced predictive power over traditional GxE models which include only a single genetic variant and a single environmental exposure. Although this approach was originally made for GxE modelling, it is flexible and does not require the use of genetic and environmental variables. It can also handle more than 2 latent variables (rather than just G and E) and 3-way interactions or more. The LEGIT model produces highly interpretable results and is very parameter-efficient thus it can even be used with small sample sizes (n < 250). Tools to determine the type of interaction (vantage sensitivity, diathesis-stress or differential susceptibility), with any number of genetic variants or environments, are available (https://psyarxiv.com/27uw8).
There are a few vignettes available to help in using the package:
- For usage of the LEGIT models, see: https://cran.r-project.org/web/packages/LEGIT/vignettes/LEGIT.html
- For usage of the GxE testing as per Belsky et al. (2013), see: https://cran.r-project.org/web/packages/LEGIT/vignettes/GxE_testing.html
- For usage of elastic net with LEGIT models, see: https://cran.r-project.org/web/packages/LEGIT/vignettes/ElasticNet.html
For the rest, please refer to the reference manual: https://cran.r-project.org/web/packages/LEGIT/LEGIT.pdf
Many are interested in using the package only for GxE testing with a standard GxE model with only one gene and one environment (so non-LEGIT). To make it work with one gene and one environment, you can simply use the GxE_interaction_test function and set genes=data.frame(G=mydata$mygene) and env=data.frame(E=mydata$myenvironment). Same goes for the LEGIT function.
The benefits of using this package for GxE testing is that it has more functionalities and output information than other SAS/SPSS software. Furthermore:
- It also tests for vantage sensitivity, while most software doesn't.
- It allows plotting the GxE easily with the plot function.
- It allows testing for rGE (gene-by-environment correlation) with the rGE function.
To install the latest stable version, run in R :
- install.packages("LEGIT")
To install the latest GitHub development version which could contain new or experimental features, run in R :
- install.packages("devtools")
- devtools::install_github("AlexiaJM/LEGIT")
Regarding the GitHub installation. When it ask "Enter one or more numbers, or an empty line to skip updates:", just write nothing and press enter. Otherwise, you risk getting a "Cannot remove prior package X" error.
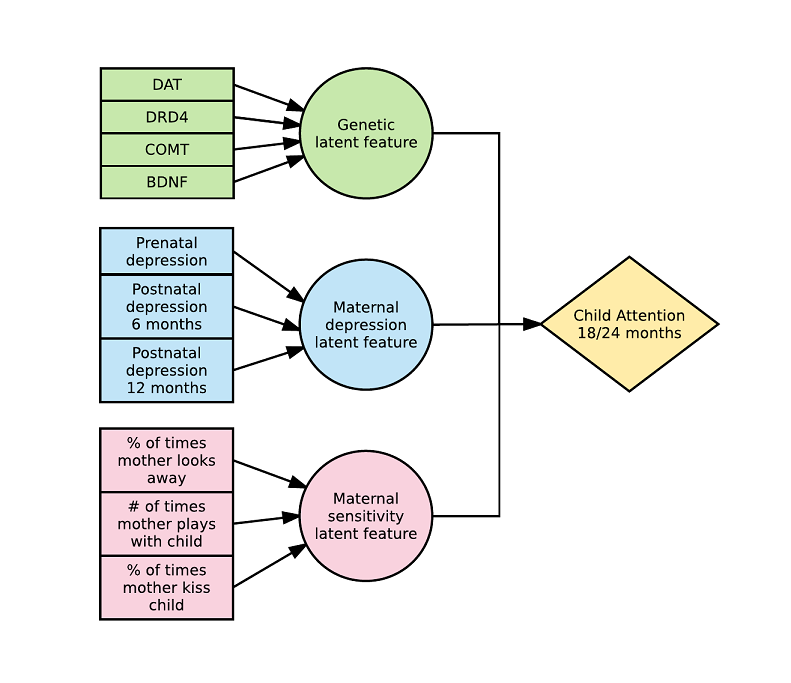
Here is an example with 2 latent variables and a 2-way interaction (see https://arxiv.org/abs/1703.08111) :
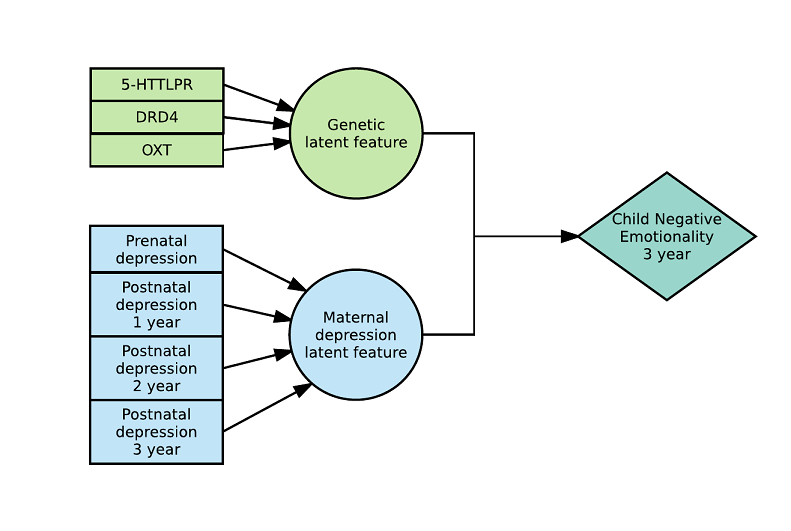
Here is an example with 3 latent variables and a 3-way interaction (see https://arxiv.org/abs/1703.08111) :