Text Algorithms is a C++ project that implements substring search and sequence alignment algorithms. This project can be useful for bioinformatics and other full-text search tasks.
The project requires the following dependencies:
- CMake >= 3.15
- C++17-compatible compiler
To build the project, follow these steps:
- Clone the repository:
git clone https://github.com/your-username/TextAlgorithms.git- Navigate to the project directory:
cd TextAlgorithms- Run the following commands:
cmake -S . -B ./build
cmake --build ./buildThe project implements the Rabin-Karp algorithm for substring search. To use it, include the SubstringSearch.h header and call the rabinKarp function with the haystack and needle strings:
#include "SubstringSearch.h"
// ...
std::string haystack = "Madam, I'm Adam";
std::string needle = "am";
std::vector<int> matches = rabinKarp(haystack, needle);
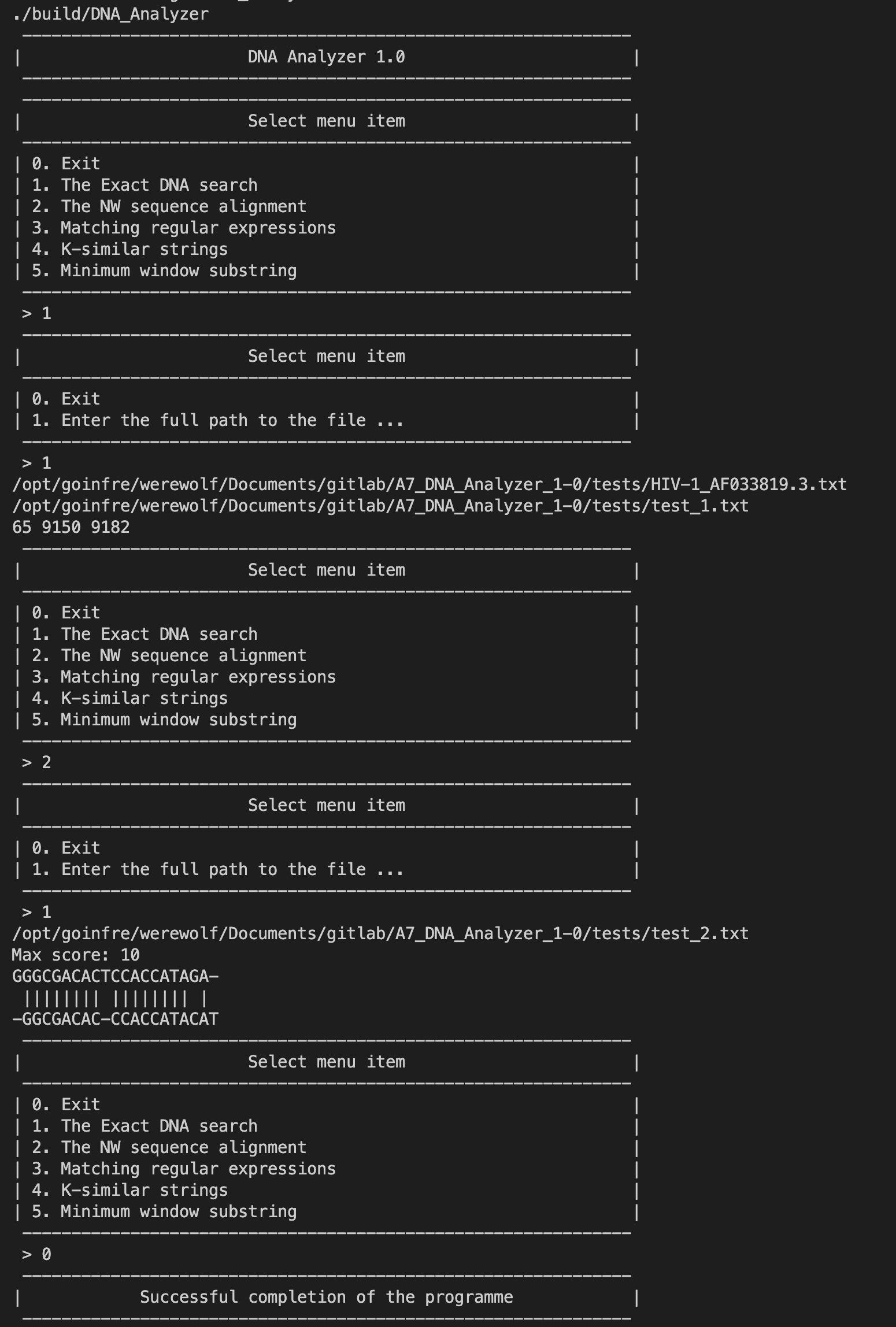
// matches contains the positions of the needle occurrences in the haystackThe project implements the Needleman-Wunsch algorithm for sequence alignment. To use it, include the SequenceAlignment.h header and call the needlemanWunsch function with the two sequences and the similarity matrix:
#include "SequenceAlignment.h"
// ...
std::string seq1 = "GGGCGACACTCCACCATAGA";
std::string seq2 = "GGCGACACCCACCATACAT";
std::vector<std::string> alignment = needlemanWunsch(seq1, seq2, similarityMatrix);
// alignment contains the two sequences aligned with gapsFind all occurrences of the string "AAGCCTCTCAAT" in the HIV virus sequence:
#include "SubstringSearch.h"
#include <fstream>
#include <iostream>
int main() {
std::ifstream file("HIV.txt");
std::string haystack((std::istreambuf_iterator<char>(file)), std::istreambuf_iterator<char>());
std::string needle = "AAGCCTCTCAAT";
std::vector<int> matches = rabinKarp(haystack, needle);
for (int match : matches) {
std::cout << "Match at position " << match << std::endl;
}
return 0;
}Align two DNA sequences using a similarity matrix:
#include "SequenceAlignment.h"
#include <iostream>
int main() {
std::string seq1 = "GGGCGACACTCCACCATAGA";
std::string seq2 = "GGCGACACCCACCATACAT";
std::vector<std::string> alignment = needlemanWunsch(seq1, seq2, similarityMatrix);
std::cout << alignment[0] << std::endl << alignment[1] << std::endl;
return 0;
}The program checks whether a sequence over the alphabet {A, C, G, T} matches a regular expression.
The input of the program is a file with two lines. The first line contains the sequence to be checked for a match. The second line contains a pattern that includes characters from the alphabet and the following characters:
.-- matches any single character from the alphabet;?-- matches any single character from the alphabet or the absence of a character;+-- matches zero or more repetitions of the previous element;*-- matches any sequence of characters from the alphabet or the absence of characters.
The output of the program is True/False - whether the given sequence matches the pattern.
Example input:
GGCGACACCCACCATACAT
G?G*AC+A*A.
Example output:
True
Strings s1 and s2 are k-similar (for some non-negative integer k) if it is possible to swap two letters in s1 exactly k times so that the resulting string is equal to s2.
The program checks k-similarity of two sequences over the alphabet {A, C, G, T}.
The input of the program is a file with two lines. The output of the program is the smallest k for which s1 and s2 are k-similar. If the strings are not anagrams, print an error message.
Example input:
GGCGACACC
AGCCGCGAC
Example output:
3
A program for finding the minimum window substring for a sequence over the alphabet {A, C, G, T}.
The input to the program is a file containing two lines: s and t. A window substring of string s is a substring that contains all characters present in string t (including duplicates).
The output of the program is the minimum length window substring. If there is no window substring, return an empty string.
Example input:
GGCGACACCCACCATACAT
TGT
Example output:
GACACCCACCATACAT
This project is licensed under the terms of the MIT license. See LICENSE for more information.