Accepted by International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2024)
Chenxin Li1, Brandon Y. Feng2✉, Yifan Liu1, Hengyu Liu1, Cheng Wang1, Weihao Yu1, Yixuan Yuan1✉
1 The Chinese University of Hong Kong, 2 Massachusetts Institute of Technology
✉ Corresponding Author.
- We present state-of-the-art results on surgical scene reconstruction from a sparse set of endoscopic views, achieving and significantly enhancing the practical usage potential of neural reconstruction methods.
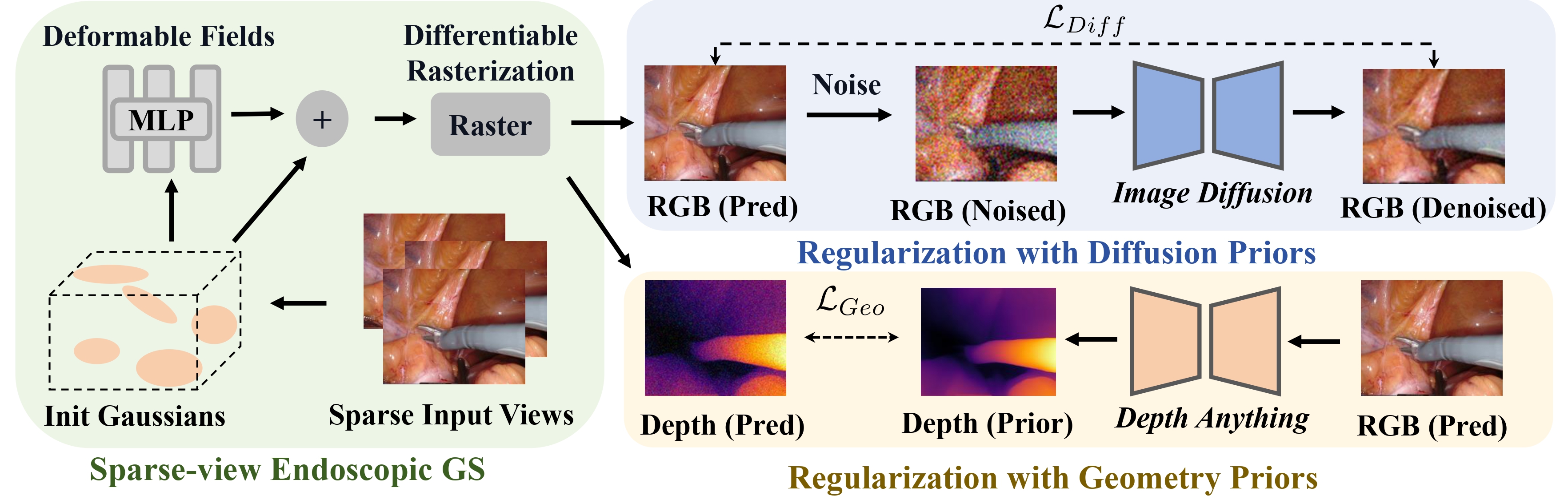
- We demonstrate an effective strategy to instill prior knowledge from a pre-trained 2D generative model to improve and regularize the visual reconstruction quality under sparse observations.
- We introduce an effective strategy to distill geometric prior knowledge from a visual foundation model that drastically improves the geometric reconstruction quality under sparse observations.
git clone https://github.com/CUHK-AIM-Group/EndoSparse.git
cd EndoSparse
conda create -n endosparse python=3.7
conda activate endosparse
pip install -r requirements.txt
pip install -e submodules/depth-diff-gaussian-rasterization
pip install -e submodules/simple-knnTips: 24 GB GPU memory is required for training and inference.
Same to the 📚Data Preparation process of EndoGaussian:
ENDONERF The dataset provided in EndoNeRF is used. You can download and process the dataset from their website. We use the two accessible clips including 'pulling_soft_tissues' and 'cutting_tissues_twice'.
SCARED The dataset provided in SCARED is used. To obtain a link to the data and code release, sign the challenge rules and email them to max.allan@intusurg.com. You will receive a temporary link to download the data and code. Follow MICCAI_challenge_preprocess to extract data. The resulted file structure is as follows.
The file structure is as follows.
├── data
│ | endonerf
│ ├── pulling
│ ├── cutting
│ | scared
│ ├── dataset_1
│ ├── keyframe_1
│ ├── data
│ ├── ...
│ ├── dataset_2
| ├── ...
Greatly appreciate the tremendous effort for the following projects!
If you find this work helpful for your project,please consider citing the following paper:
@article{li2024endosparse,
author = {Chenxin Li and Brandon Y. Feng and Yifan Liu and Hengyu Liu and Cheng Wang and Weihao Yu and Yixuan Yuan},
title = {EndoSparse: Real-Time Sparse View Synthesis of Endoscopic Scenes using Gaussian Splatting},
journal = {arXiv preprint},
year = {2024}
}