Accepted by International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2024 poster)
Hengyu Liu*, Yifan Liu*, Chenxin Li*, Wuyang Li, Yixuan Yuan✉
The Chinese University of Hong Kong
* Equal Contribution, ✉ Corresponding Author.
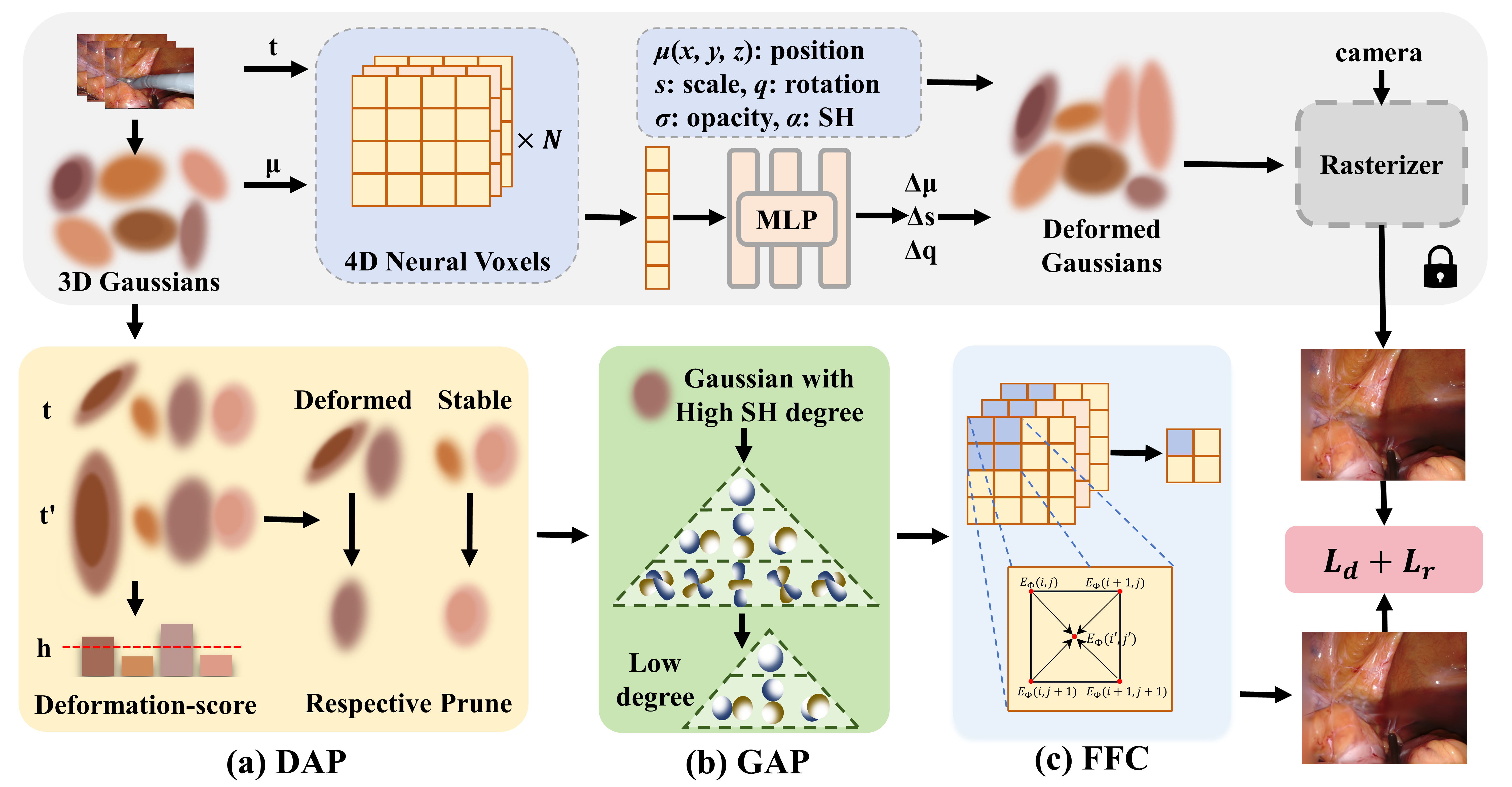
- We propose a holistic Lightweight 4D Gaussian Splatting (LGS) framework that allows for achieving satisfactory endoscopic reconstruction with both efficient rendering and storing.
- We present a Deformation-Aware Pruning (DAP) which alleviates the Quantity burden of Gaussian representation.
- We propose a Gaussian-Attribute Pruning (GAP), which addresses the High-dimension burden of Gaussian attributes.
- We present Feature Field Condensation (FFC) which mitigates the High-resolution burden of spatial-temporal deformable fields.
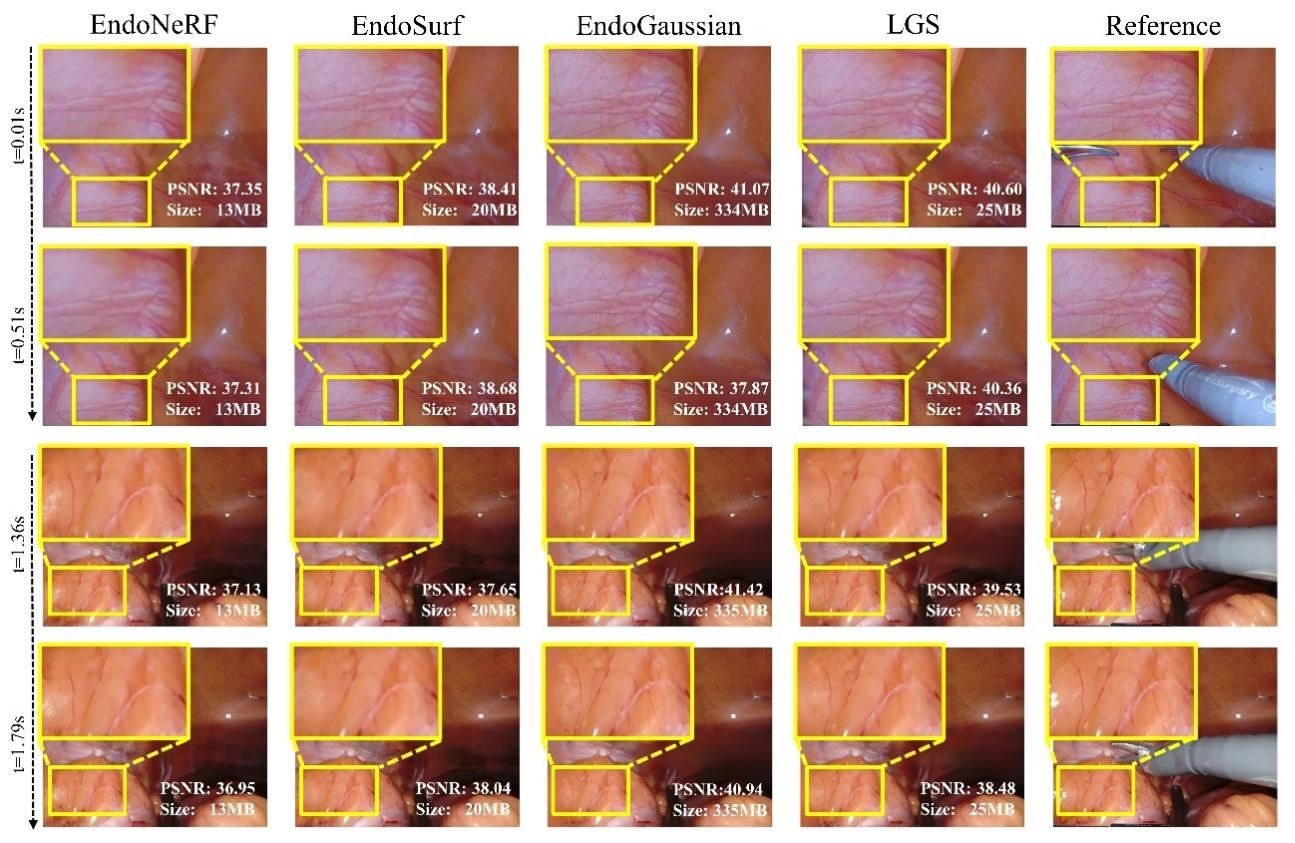
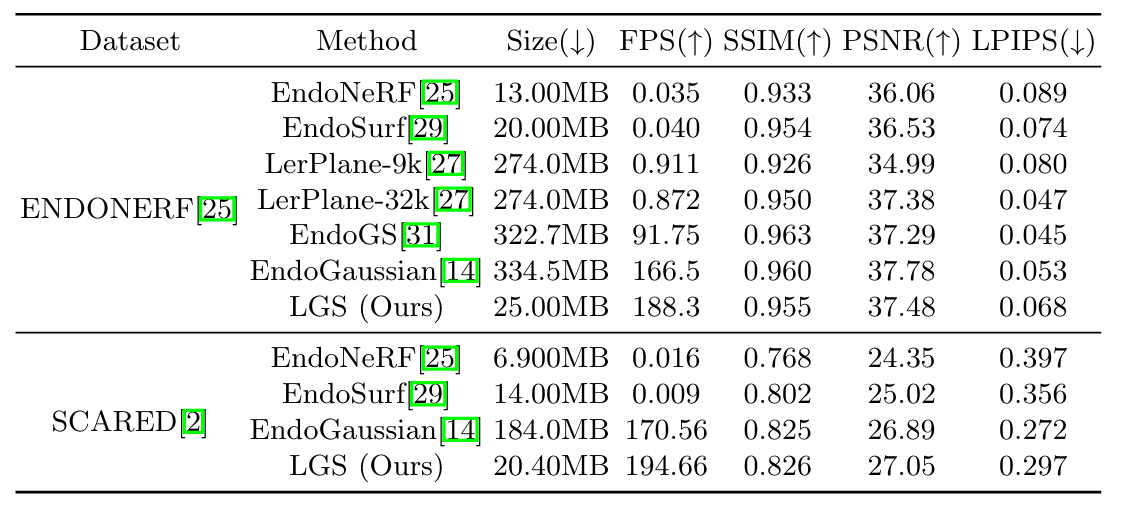
- Experimental results show that LGS can achieve higher storage efficiency with an over
$9\times$ compression rate, whilst maintaining pleasing reconstruction quality and rendering speed.
submodules/simple_knncan be download from herecompress-diff-gaussian-rasterizationcan be download from compress-diff-gaussian-rasterization
git clone https://github.com/CUHK-AIM-Group/LGS.git
cd LGS
conda create -n lgs python=3.7
conda activate lgs
pip install -r requirements.txt
pip install -e submodules/depth-diff-gaussian-rasterization
pip install -e submodules/simple-knnTips: 2 GB GPU memory is required for training and inference.
Same to the 📚Data Preparation process of EndoGaussian:
ENDONERF The dataset provided in EndoNeRF is used. You can download and process the dataset from their website. We use the two accessible clips including 'pulling_soft_tissues' and 'cutting_tissues_twice'.
SCARED The dataset provided in SCARED is used. To obtain a link to the data and code release, sign the challenge rules and email them to max.allan@intusurg.com. You will receive a temporary link to download the data and code. Follow MICCAI_challenge_preprocess to extract data. The resulted file structure is as follows.
The file structure is as follows.
├── data
│ | endonerf
│ ├── pulling
│ ├── cutting
│ | scared
│ ├── dataset_1
│ ├── keyframe_1
│ ├── data
│ ├── ...
│ ├── dataset_2
| ├── ...
Run the following code to train LGS in a distillation scheme.
cd to this repo
conda activate engs
dataset="name/of/dataset"
test_name="name/of/output/file"
config_name="name/of/config"
check_point="/path/to/teacher/model/chkpntN.pth"
deformation_path="/path/to/teacher/model/point_cloud/iteration_N"
CUDA_VISIBLE_DEVICES=GPU_ID python train.py \
-s /path/to/endonerf/cutting_tissues_twice/ \
-m /path/to/endonerf/endonerf-output/ \
--expname $dataset/new_init/$test_name \
--start_checkpoint $check_point \
--deformatioin_model_path $deformation_path \
--teacher_model $check_point \
--enable_covariance \
--new_max_sh 2 \
--iteration 4000 \
--configs $config_name \
--prune_percent 0.3 \
--prune_decay 0.6 \
--prune_iterations 2000 \
--prune \
--prune_threshold 0.5 \
--prune_sh \
--prune_deform \
--v_pow 0.1 \
--gt \
--port $port > /path/to/log
Notice:
- (1) Teacher Model is a trained EndoGaussian model, while the size of the deformation filed of teacher model should be set the same as LGS:
kplanes_config = {
'grid_dimensions': 2,
'input_coordinate_dim': 4,
'output_coordinate_dim': 64,
'resolution': [64, 64, 64, 100]
}
- (2) While training EndoGaussian, EndoGaussian's two saving processs are at different times, one before pruning the Gaussians and the other after pruning. Please refer to here and get the consistent models.
Run the following code to render and get the evaluaton.
CUDA_VISIBLE_DEVICES=GPU_ID python render.py --model_path /path/to/trained/model \
--skip_train \
--new_max_sh 2 \
--prune_deform \
--configs $config_name > /path/to/log
CUDA_VISIBLE_DEVICES=GPU_ID python metrics.py --model_path /path/to/trained/model > /path/to/log- Release code for LGS
- Clean up the code for LGS
- Updae the code for ablation experiments
Greatly appreciate the tremendous effort for the following projects!
If you find this work helpful for your project,please consider citing the following paper:
@article{liu2024lgs,
title={LGS: A Light-weight 4D Gaussian Splatting for Efficient Surgical Scene Reconstruction},
author={Liu, Hengyu and Liu, Yifan and Li, Chenxin and Li, Wuyang and Yuan, Yixuan},
journal={arXiv preprint arXiv:2406.16073},
year={2024}
}