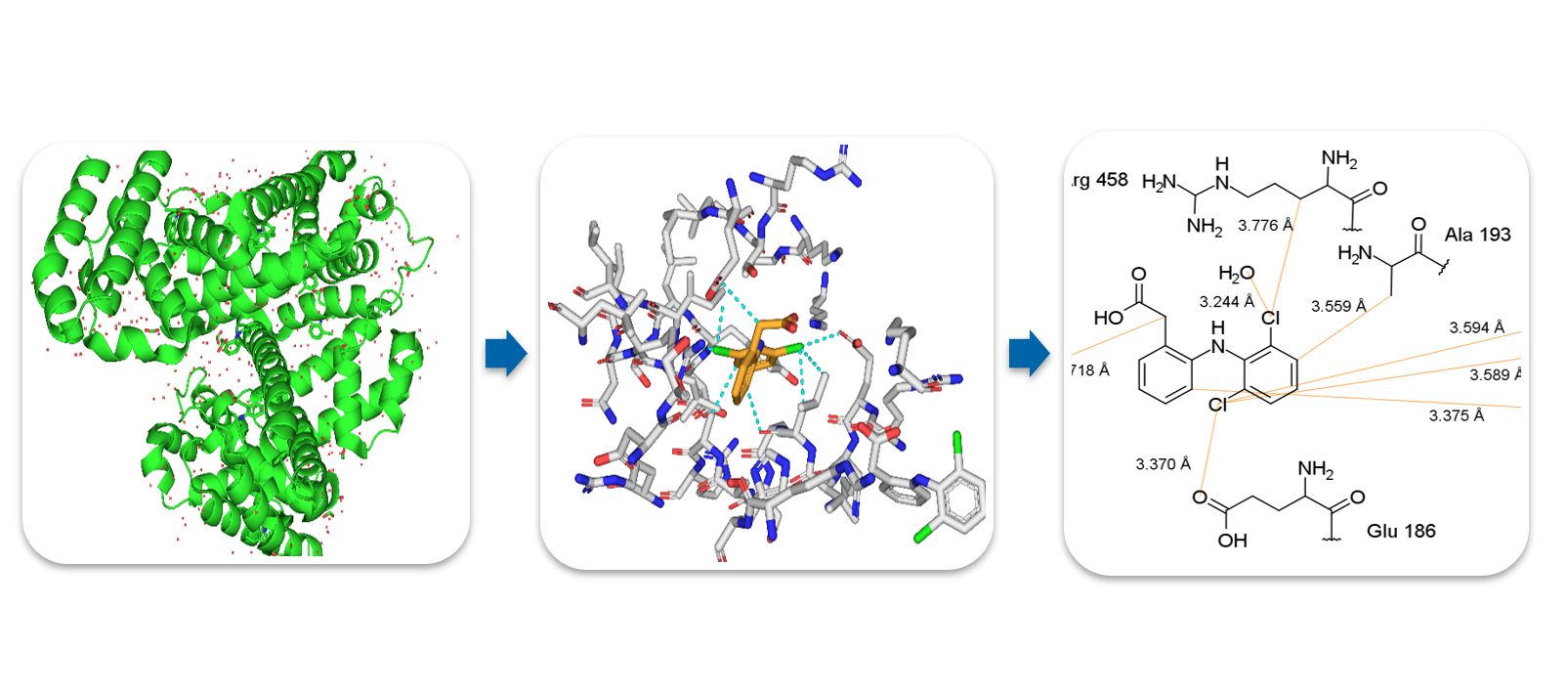
A program analyzing 3D protein structures from PDB to generate 2D binding motifs. The current version creates .mrv-files that can be open with MarvinSketch for example. The .mrv-file shows interactions between ligand and protein atoms.
This program is tested and developed on a Windows platform, so some platform related errors may occur if you don't use a Windows platform.
To ensure optimal workflow first install Python from here (I use v3.7.3). Then get Anaconda from here. After you've installed Anaconda open Anaconda Prompt and type
conda install -c schrodinger pymol.
Next install the required packages using
pip install pysmiles
and
pip install tabulate.
To view the generated .mrv-file (xml like) I recommend using MarvinSketch (download from here). To use this you have to create an account and sign in.
To get the latest version of StructureAnalyzer use
git clone https://github.com/Cardypro/StructureAnalyzer.git
After downloading StructureAnalyzer open Anaconda Prompt, navigate to the path where you installed the StructureAnalyzer and execute
pymol StructureAnalyzer.py
Pymol should start now. Type
StructureAnalyzer("PDB-code", "Ligand code", "condition", ignoreH2O, defaultRadius)
e.g.
StructureAnalyzer("6hn0", "DIF", "N 1.1*vdw O|N", True)
to analyze the interactions of the PDB entry named 6HN0 with Diclofenac within a cutoff range 1.1 times of the van-der-Waals distance around the Diclofenac. It's also restricted to ligands-Nitrogen and pocket-Oxygen or -Nitrogen.
The produced .mrv-file can be found in the /Output directory. You can open the file using MarvinSketch. The interactions are represented by orange lines by default. The names of the residues are connected to the corresponding residue by a thin grey line so you can easily rearange them in an aesthetical way.
The program is optimized and mainly tested for 6HN0 and DIF. Although it will (hopefully) work for (almost) all codes given in the PDB. Arguments marked by an asterisk * are mandatory.
The PDB-code is the four - letter - figure - code given by the PDB specifying the protein-ligand-interaction you want to investigate. The program automatically downloads the file and saves it as .cif file in your working directory.
The ligand code is the three - letter - figure - code given by the PDB specifying the ligand you want to investigate. If there are more than one protein-ligand-interactions (e.g. because there are more than one molecule of Ligand in the structure) the program will select the innermost ligand. This ensures that there are no interactions ignored because of missing information (no ligands on the edge of your structure are selected because some interactions may be missing in the file). This selection is sensitive to the geometry, not to the mass corrected centre (centre of geometry is used instead of centre of mass).
The condition determines which interactions are depicted. It always insists of three statements separated by whitespaces.
The first and the third statement determines which elements are allowed on the ligand side and the pocket/protein side repectively. Multiple elements can be allowed by separating them with a "|" (pipe). "*" (asterisk) means "all elements". The currently supported elements can be found below.
van der Waals radii
"H": 1.10,
"Li": 1.81,
"Na": 2.27,
"K": 2.75,
"Rb": 3.03,
"Cs": 3.43,
"Fr": 3.48, #End I
"Be": 1.53,
"Mg": 1.73,
"Ca": 2.31,
"Sr": 2.49,
"Ba": 2.68,
"Ra": 2.83, #End II
"B": 1.92,
"Al": 1.84,
"Ga": 1.87,
"In": 1.93,
"Tl": 1.96, #End III
"C": 1.70,
"Si": 2.10,
"Ge": 2.11,
"Sn": 2.17,
"Pb": 2.02, #End IV
"N": 1.55,
"P": 1.80,
"As": 1.85,
"Sb": 2.06,
"Bi": 2.07, #End V
"O": 1.52,
"S": 1.80,
"Se": 1.90,
"Te": 2.06,
"Po": 1.97, #End VI
"F": 1.47,
"Cl": 1.75,
"Br": 1.83,
"I": 1.98,
"At": 2.02, #End VII
"He": 1.40,
"Ne": 1.54,
"Ar": 1.88,
"Kr": 2.02,
"Xe": 2.16,
"Rn":2.20 #End Main Group
The middle statement determines the cutoff. There are two ways to use this. First, you can just type a float representing a constant cutoff for interactions.
The second way is to determine the cutoff on a dynamical way. Therefore the van-der-Waals-radii are used. The vdw-radii are currently obtained from [1]. The cutoff is then calculated as the sum of the van-der-Waals-radii of the interacting elements (multiplied by an optional factor). To use the second way your second statement must be something like "factor*vdw".
The cutoff is calculated by using simple 3D geometry (Pythagorean theorem). The program does not evaluate whether the found interactions make sense in a chemical view. The default condition is "* 1*vdw *", analyzing all atoms within a cutoff of the sum of the vdw-radii.
This decides whether to ignore water molecules or not. The default value is False so water molecules are depicted.
The fallback radius that should be used if the radius for the current element is not defined in the vdwRadii-table (see above). If there is no default radius given, not lited elements won't be evaluated.
Determines the size of the pocket that is shown in pyMOL. The default value is 8 A.
Determines if the interaction table (below) should be written in a markdown (github format) file. Default is True.
Example of an interaction table
| atom ligand | distance [A] | atom pocket | element |
|---|---|---|---|
| DIF 605/CL2 | 3.27785 | ARG 208/CG | C |
| DIF 605/C1 | 3.15951 | HOH 1016/O | O |
| DIF 605/N1 | 2.82841 | HOH 884/O | O |
| DIF 605/O1 | 3.10756 | DIF 606/C5 | C |
| DIF 605/O2 | 2.71713 | ASP 323/O | O |
| DIF 605/O2 | 3.09461 | DIF 606/C5 | C |
| DIF 605/O2 | 3.08978 | DIF 606/C6 | C |
| DIF 605/C8 | 2.93876 | HOH 884/O | O |
| DIF 605/C14 | 3.03511 | DIF 606/C5 | C |
| atom ligand | distance [A] | atom pocket | element |
|---|---|---|---|
| DIF 606/C5 | 3.10756 | DIF 605/O1 | O |
| DIF 606/C5 | 3.09461 | DIF 605/O2 | O |
| DIF 606/C5 | 3.03511 | DIF 605/C14 | C |
| DIF 606/C6 | 3.08978 | DIF 605/O2 | O |
| DIF 606/C11 | 3.10796 | GLU 353/OE2 | O |
| DIF 606/C11 | 2.93863 | HOH 815/O | O |
| DIF 606/C12 | 2.99284 | HOH 815/O | O |
Instead of using the StructureAnalyzer command you can use the multipleAnalyzer. This allows you to analyze more than one pdb-code at once. It works similar to the StructureAnalyzer except it takes an array of strings containing the protein codes to be analyzed.
E.g. multipleAnalyzer(["6hn0","6hn1"], "DIF")
If you get errors like
An unexpected error has occurred. Conda has prepared the above report.
make sure you've installed Pymol after you've installed Anaconda.
This program uses pysmiles writen by Peter C. Kroon - special thanks to him. The program generates network graphs obtained by Depth-first searching the ligand and generating a graph with atoms as nodes and bonds as edges. Since PyMOL isn't able to give information about the bond order PySmiles calculates the bond order from one atom to another.
- In future versions there should be some more aesthetic options like different colours for different types of interactions.
- There are some issues evaluating hydrogen bonds that has to be investigated. For example, it's not quite sure how the hydrogen atom has to be taken into account.
- In future versions other non-pdb-files should also be evaluable.
[1] M. Mantina, A. C. Chamberlin, R. Valero, C. J. Cramer, D. G. Truhlar, J. Phys. Chem. A 2009, 113, 5806, doi: 10.1021/jp8111556.
- The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.
- P. C. Kroon, pySmiles v1.0.0 (2018), https://github.com/pckroon/pysmiles.
StructureAnalyzer and PySmiles are distributed under the Apache 2.0 license.
Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with the License. You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.