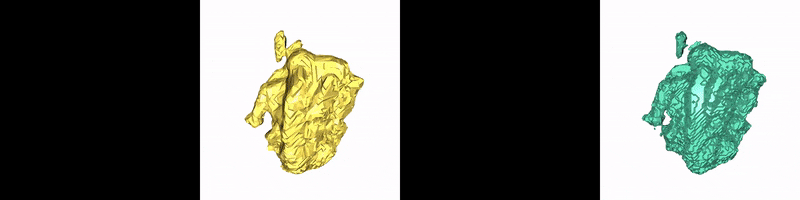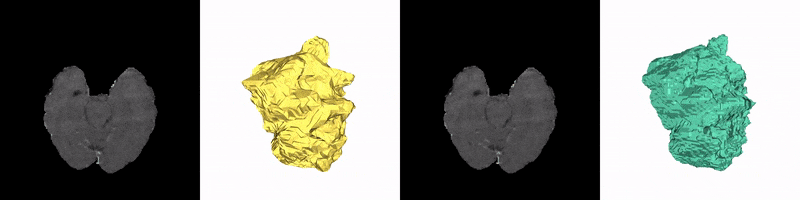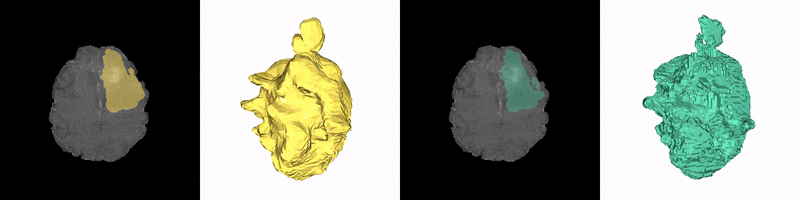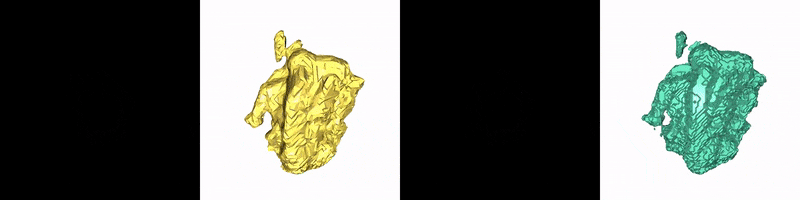
Christoph Reich, Tim Prangemeier, Özdemir Cetin & Heinz Koeppl
| Project Page | Paper | Slides | Video |
This repository includes the official and maintained PyTorch implementation of the paper OSS-Net: Memory Efficient High Resolution Semantic Segmentation of 3D Medical Data.
Convolutional neural networks (CNNs) are the current state-of-the-art meta-algorithm for volumetric segmentation of medical data, for example, to localize COVID-19 infected tissue on computer tomography scans or the detection of tumour volumes in magnetic resonance imaging. A key limitation of 3D CNNs on voxelised data is that the memory consumption grows cubically with the training data resolution. Occupancy networks (O-Nets) are an alternative for which the data is represented continuously in a function space and 3D shapes are learned as a continuous decision boundary. While O-Nets are significantly more memory efficient than 3D CNNs, they are limited to simple shapes, are relatively slow at inference, and have not yet been adapted for 3D semantic segmentation of medical data. Here, we propose Occupancy Networks for Semantic Segmentation (OSS-Nets) to accurately and memory-efficiently segment 3D medical data. We build upon the original O-Net with modifications for increased expressiveness leading to improved segmentation performance comparable to 3D CNNs, as well as modifications for faster inference. We leverage local observations to represent complex shapes and prior encoder predictions to expedite inference. We showcase OSS-Net's performance on 3D brain tumour and liver segmentation against a function space baseline (O-Net), a performance baseline (3D residual U-Net), and an efficiency baseline (2D residual U-Net). OSS-Net yields segmentation results similar to the performance baseline and superior to the function space and efficiency baselines. In terms of memory efficiency, OSS-Net consumes comparable amounts of memory as the function space baseline, somewhat more memory than the efficiency baseline and significantly less than the performance baseline. As such, OSS-Net enables memory-efficient and accurate 3D semantic segmentation that can scale to high resolutions.
If you find this research useful in your work, please cite our paper:
@inproceedings{Reich2021,
title={{OSS-Net: Memory Efficient High Resolution Semantic Segmentation of 3D Medical Data}},
author={Reich, Christoph and Prangemeier, Tim and Cetin, {\"O}zdemir and Koeppl, Heinz},
booktitle={British Machine Vision Conference},
year={2021},
organization={British Machine Vision Association},
}All required Python packages can be installed by:
pip install -r requirements.txtTo install the official implementation of the Padé Activation Unit [1] (taken from the official repository) run:
cd pade_activation_unit/cuda
python setup.py build installThe code is tested with PyTorch 1.8.1 and CUDA 11.1 on Linux with Python 3.8.5! Using other PyTorch and CUDA versions newer than PyTorch 1.7.0 and CUDA 10.1 should also be possible.
The BraTS 2020 dataset can be downloaded here and the LiTS dataset can be downloaded here. Please note, that accounts are required to login and downlaod the data on both websites.
The used training and validation split of the BraTS 2020 dataset is available here.
For generating the border maps, necessary if border based sampling is utilized, please use the
generate_borders_bra_ts_2020.py and generate_borders_lits.py script.
Table 1. Segmentation results of trained networks. Weights are generally available here and specific models are linked below.
| Model | Dice ( |
IoU ( |
Dice ( |
IoU ( |
||
|---|---|---|---|---|---|---|
| O-Net [2] | 0.7016 | 0.5615 | 0.6506 | 0.4842 | - | - |
| OSS-Net A | 0.8592 | 0.7644 | 0.7127 | 0.5579 | weights BraTS | weights LiTS |
| OSS-Net B | 0.8541 | 0.7572 | 0.7585 | 0.6154 | weights BraTS | weights LiTS |
| OSS-Net C | 0.8842 | 0.7991 | 0.7616 | 0.6201 | weights BraTS | weights LiTS |
| OSS-Net D | 0.8774 | 0.7876 | 0.7566 | 0.6150 | weights BraTS | weights LiTS |
To reproduce the results presented in Table 1, we provide multiple sh scripts, which can be found in the scripts folder. Please change the dataset path and CUDA devices according to your system.
To perform training runs with different settings use the command line arguments of the train_oss_net.py file.
The train_oss_net.py takes the following command line arguments:
| Argument | Default value | Info |
|---|---|---|
| --train | False |
Binary flag. If set training will be performed. |
| --test | False |
Binary flag. If set testing will be performed. |
| --cuda_devices | "0, 1" |
String of cuda device indexes to be used. Indexes must be separated by a comma. |
| --cpu | False |
Binary flag. If set all operations are performed on the CPU. (not recommended) |
| --epochs | 50 |
Number of epochs to perform while training. |
| --batch_size | 8 |
Number of epochs to perform while training. |
| --training_samples | 2 ** 14 |
Number of coordinates to be samples during training. |
| --load_model | "" |
Path to model to be loaded. |
| --segmentation_loss_factor | 0.1 |
Auxiliary segmentation loss factor to be utilized. |
| --network_config | "" |
Type of network configuration to be utilized (see). |
| --dataset | "BraTS" |
Dataset to be utilized. ("BraTS" or "LITS") |
| --dataset_path | "BraTS2020" |
Path to dataset. |
| --uniform_sampling | False |
Binary flag. If set locations are sampled uniformly during training. |
Please note that the naming of the different OSS-Net variants differs in the code between the paper and Table 1.
To perform inference, use the inference_oss_net.py script. The script takes the following command line arguments:
| Argument | Default value | Info |
|---|---|---|
| --cuda_devices | "0, 1" |
String of cuda device indexes to be used. Indexes must be separated by a comma. |
| --cpu | False |
Binary flag. If set all operations are performed on the CPU. (not recommended) |
| --load_model | "" |
Path to model to be loaded. |
| --network_config | "" |
Type of network configuration to be utilized (see). |
| --dataset | "BraTS" |
Dataset to be utilized. ("BraTS" or "LITS") |
| --dataset_path | "BraTS2020" |
Path to dataset. |
During inference the predicted occupancy voxel grid, the mesh prediction, and the label as a mesh are saved. The meshes are saved as PyTorch (.pt) files and also as .obj files. The occupancy grid is only saved as a PyTorch file.
We thank Marius Memmel and Nicolas Wagner for the insightful discussions, Alexander Christ and Tim Kircher for giving feedback on the first draft, and Markus Baier as well as Bastian Alt for aid with the computational setup.
This work was supported by the Landesoffensive für wissenschaftliche Exzellenz as part of the LOEWE Schwerpunkt CompuGene. H.K. acknowledges support from the European Re- search Council (ERC) with the consolidator grant CONSYN (nr. 773196). O.C. is supported by the Alexander von Humboldt Foundation Philipp Schwartz Initiative.
[1] @inproceedings{Molina2020Padé,
title={{Pad\'{e} Activation Units: End-to-end Learning of Flexible Activation Functions in Deep Networks}},
author={Alejandro Molina and Patrick Schramowski and Kristian Kersting},
booktitle={International Conference on Learning Representations},
year={2020}
}[2] @inproceedings{Mescheder2019,
title={{Occupancy Networks: Learning 3D Reconstruction in Function Space}},
author={Mescheder, Lars and Oechsle, Michael and Niemeyer, Michael and Nowozin, Sebastian and Geiger, Andreas},
booktitle={CVPR},
pages={4460--4470},
year={2019}
}