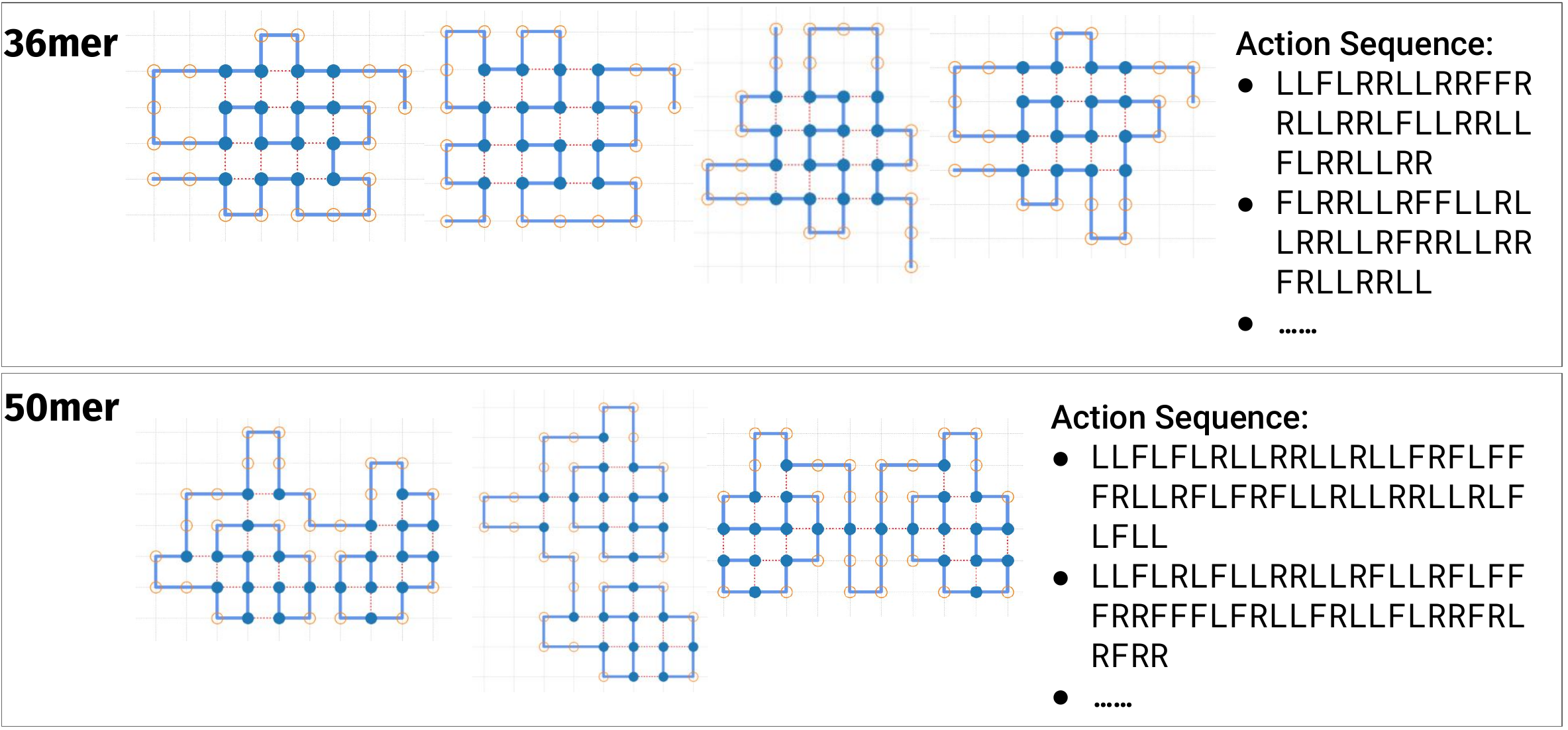
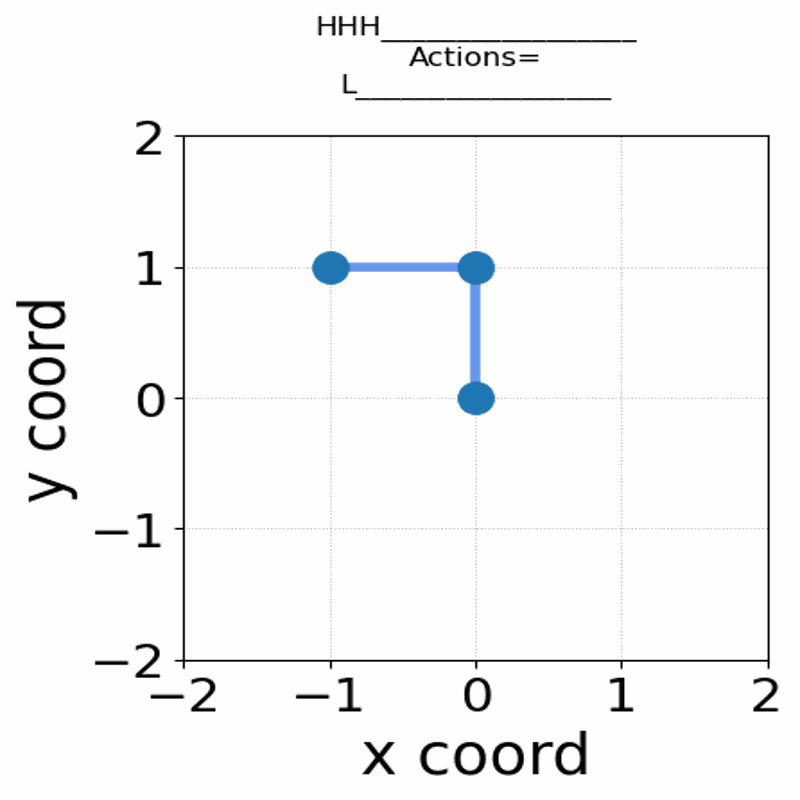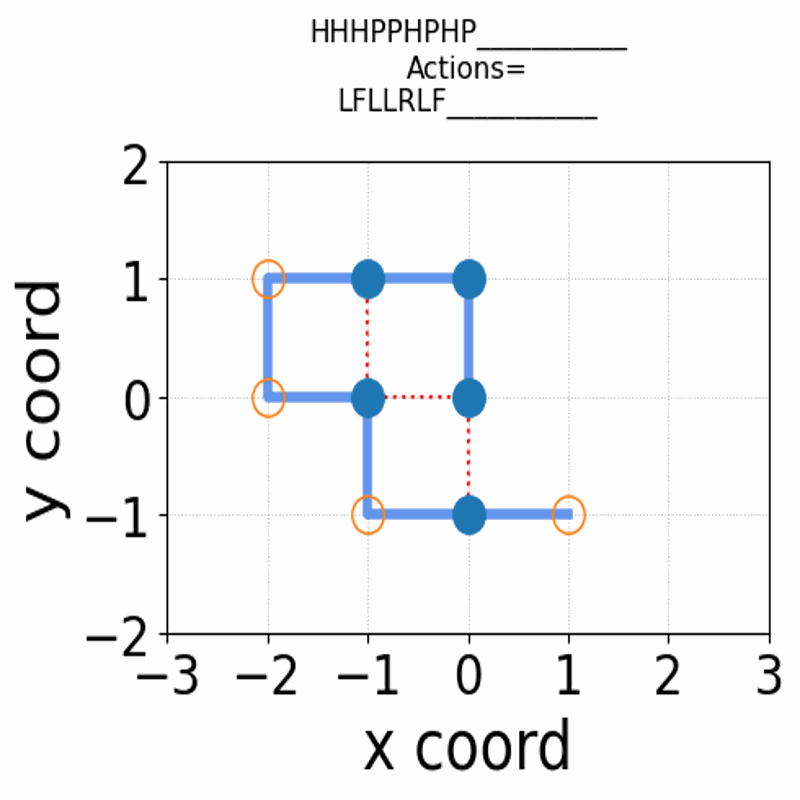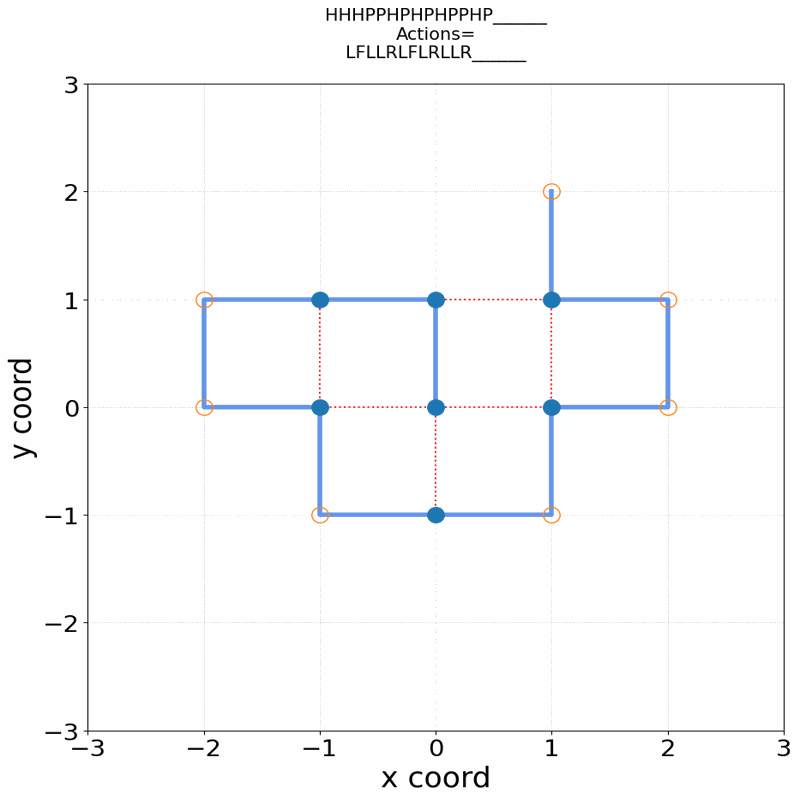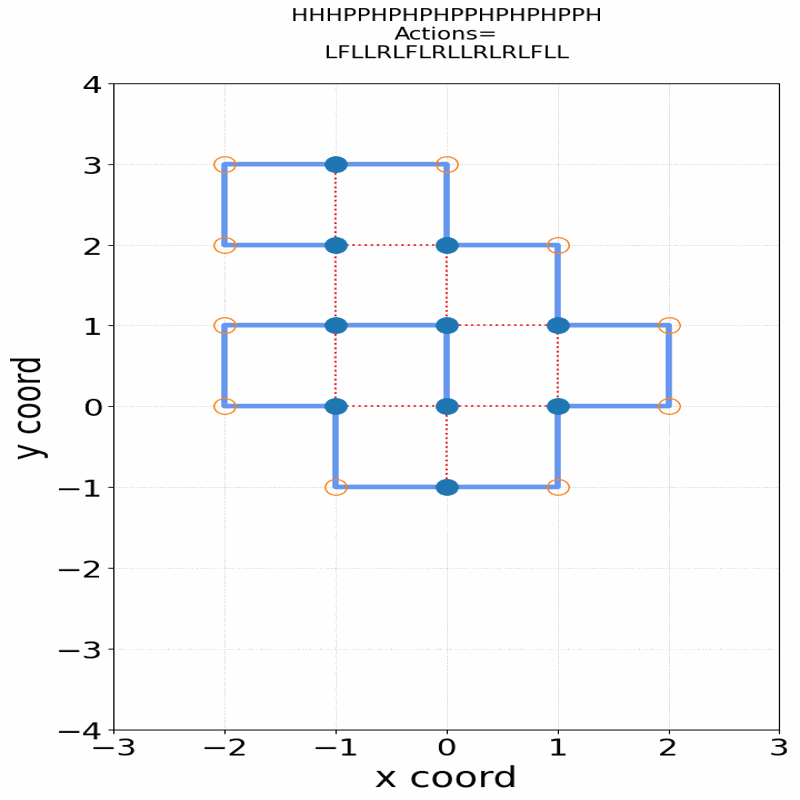
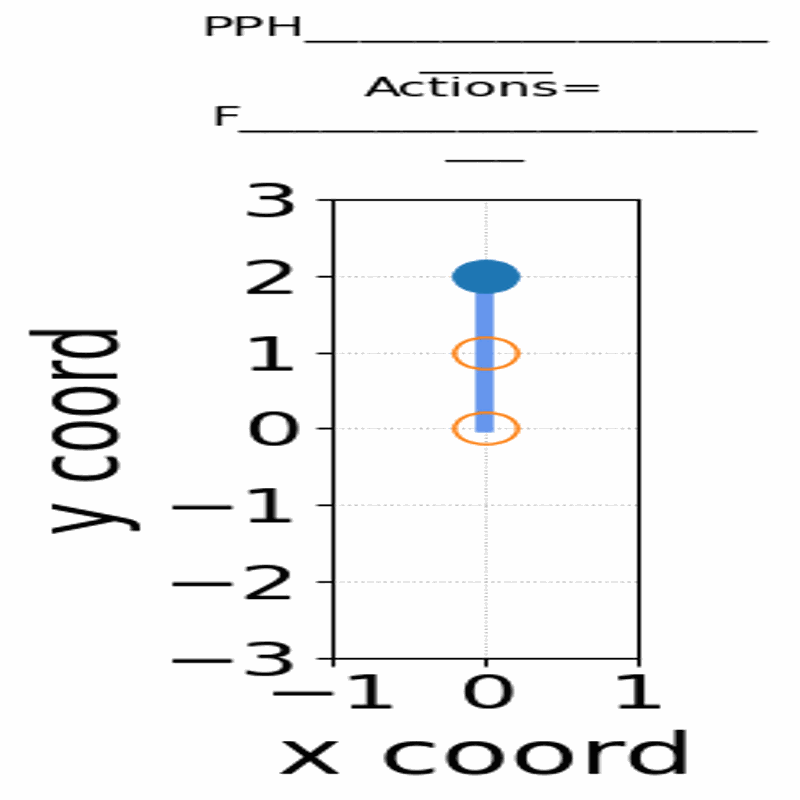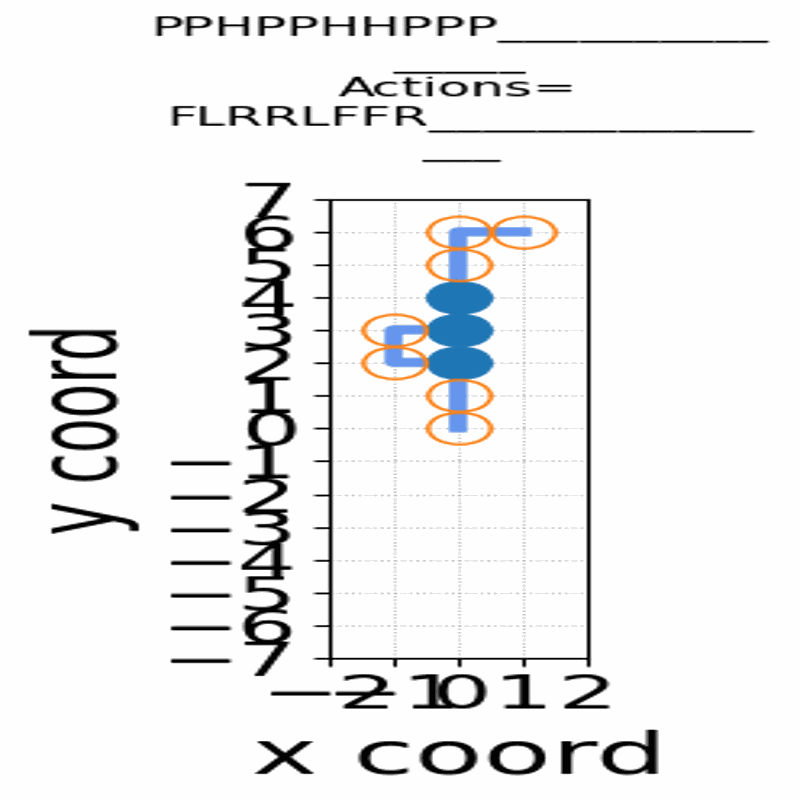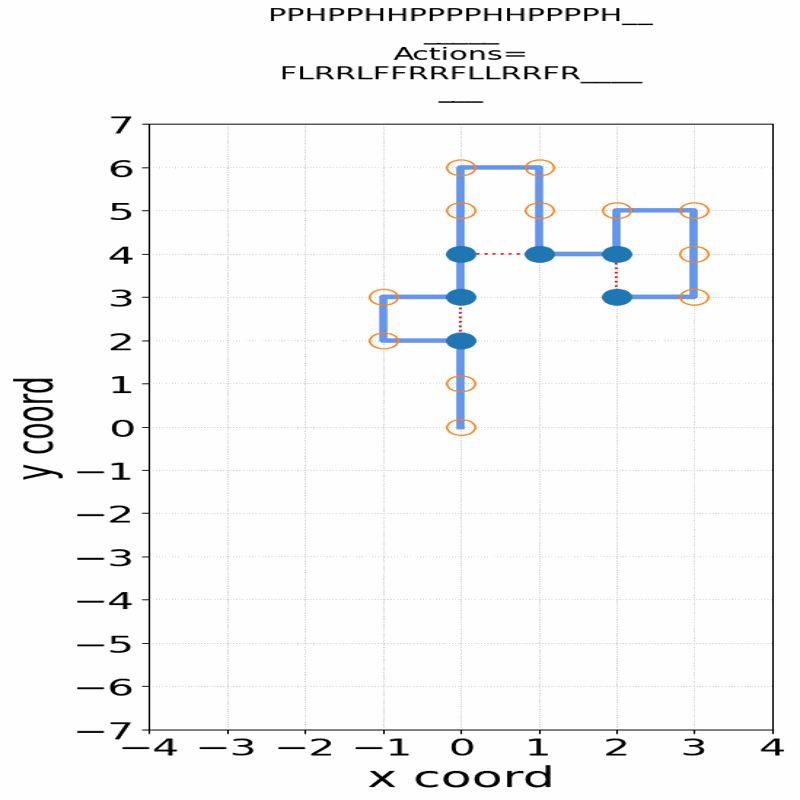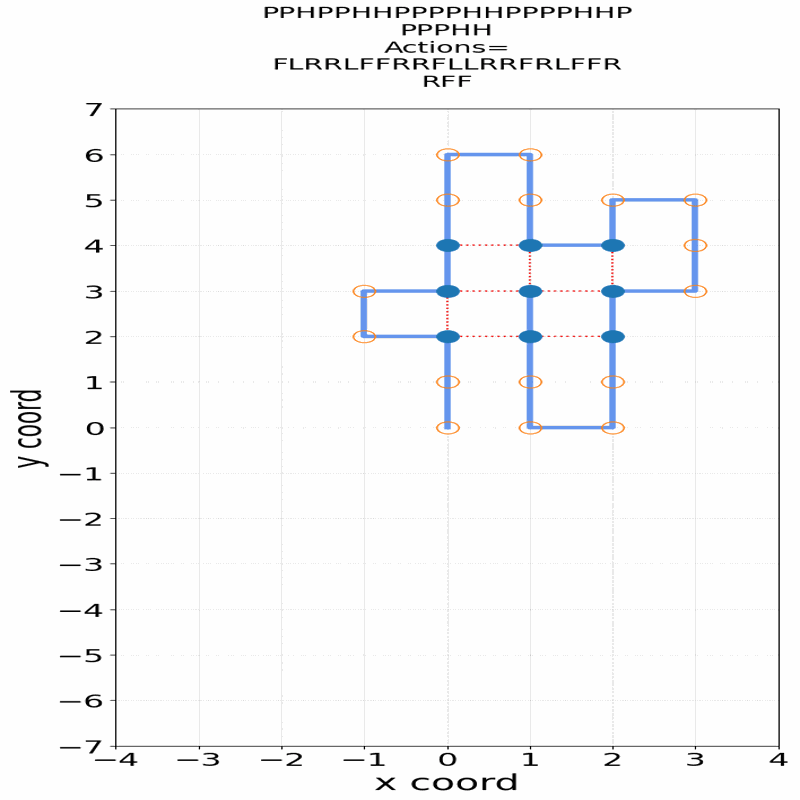
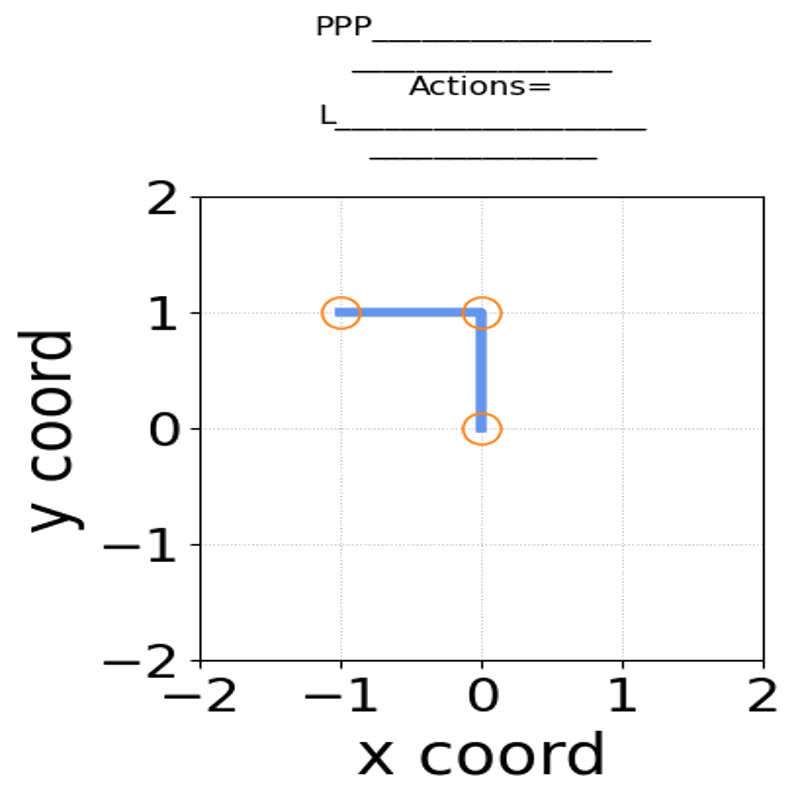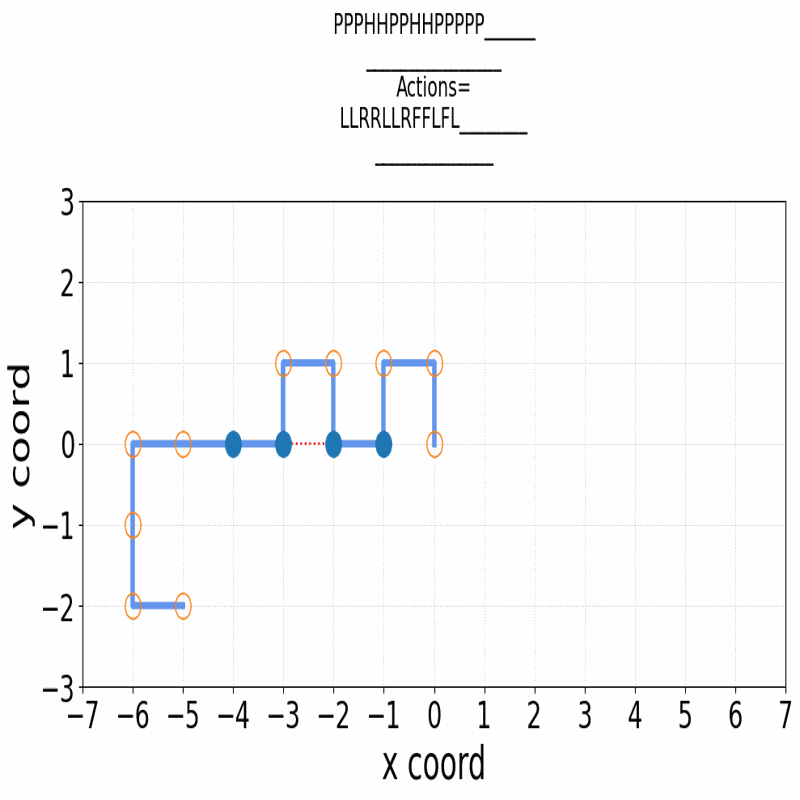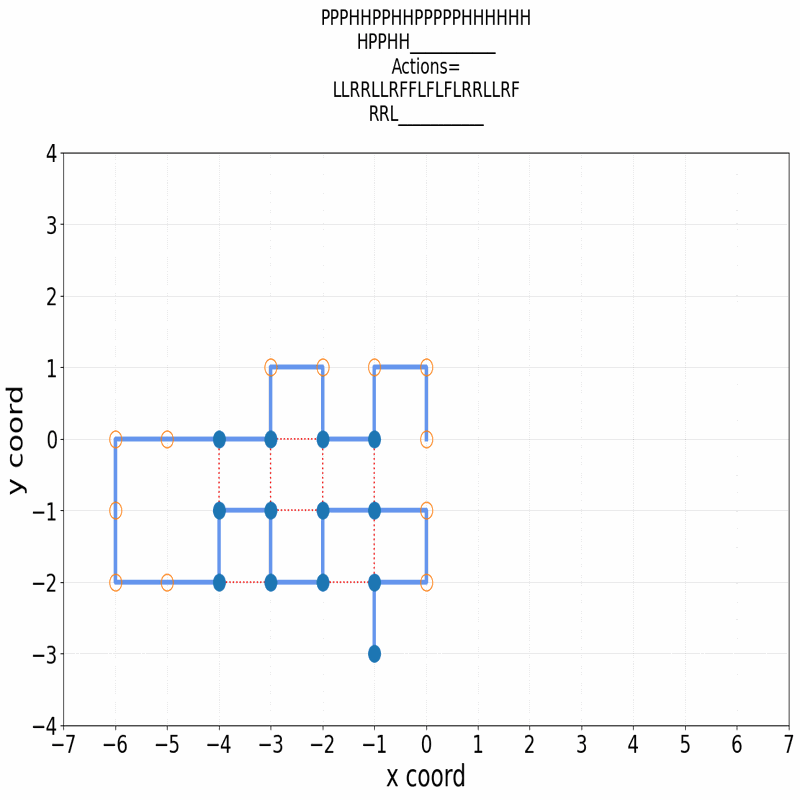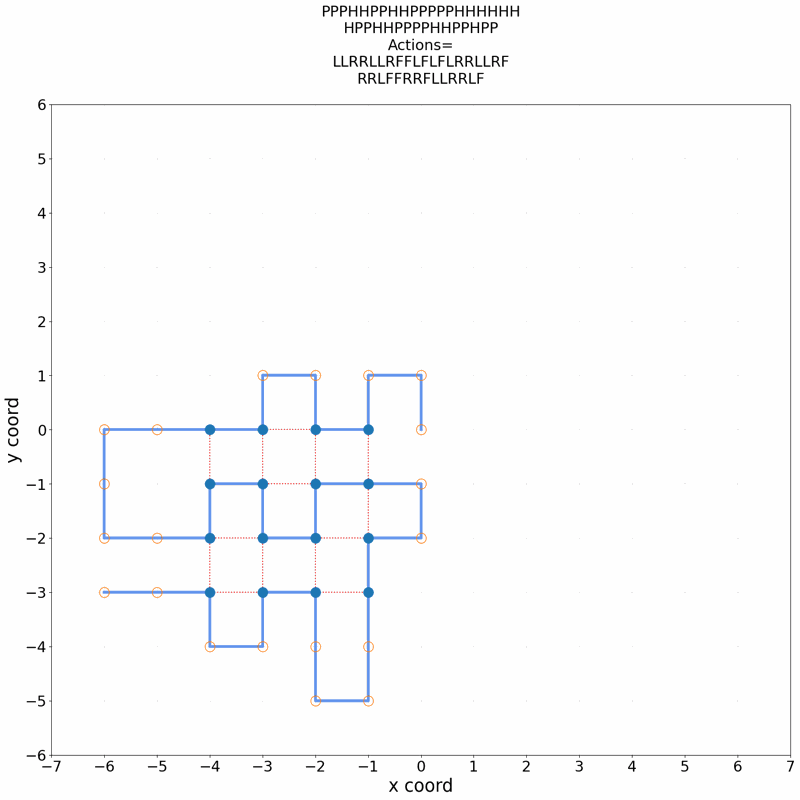
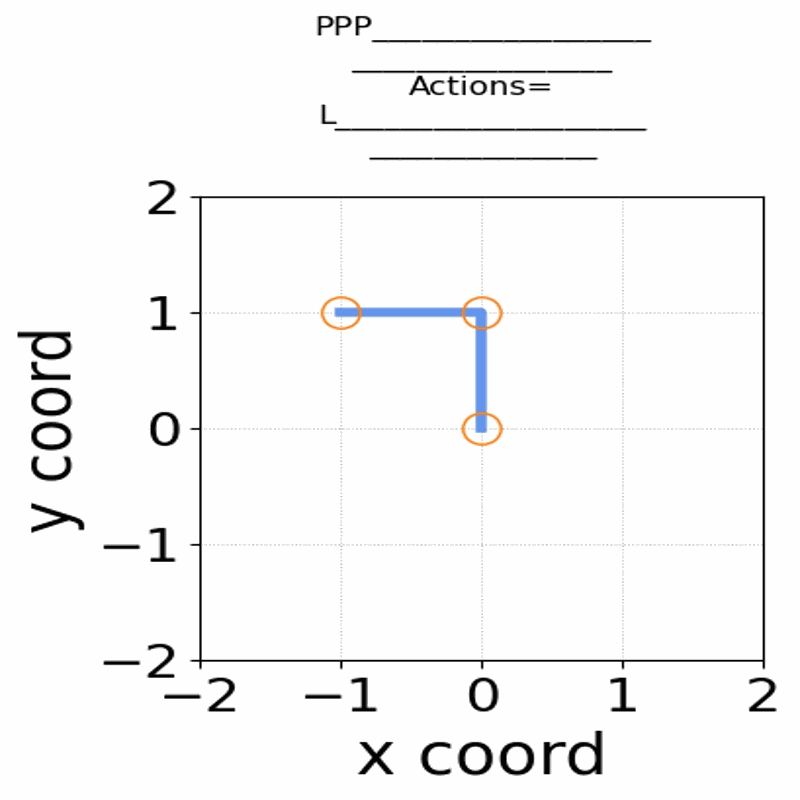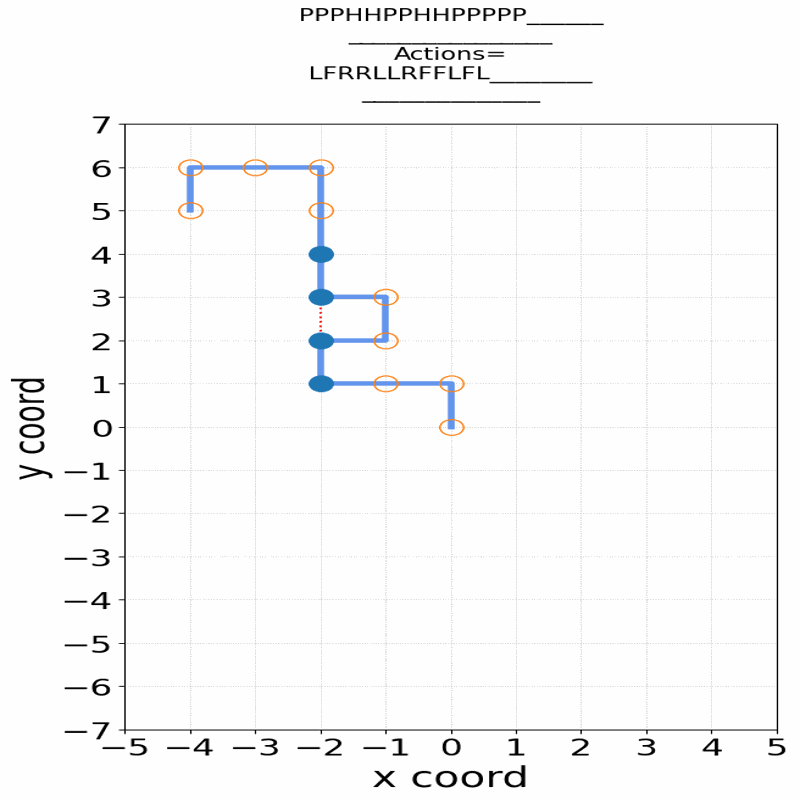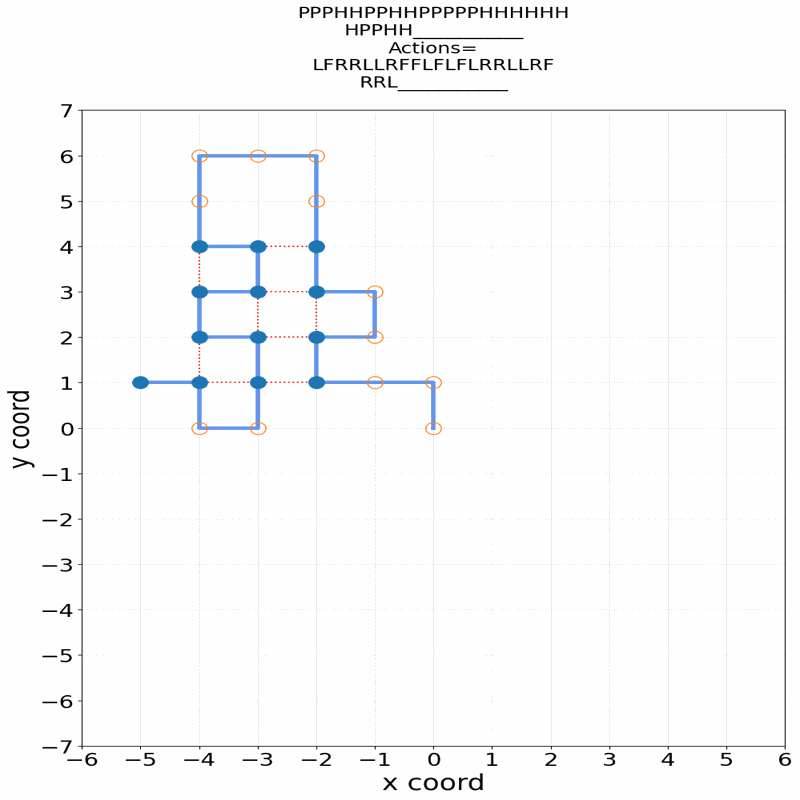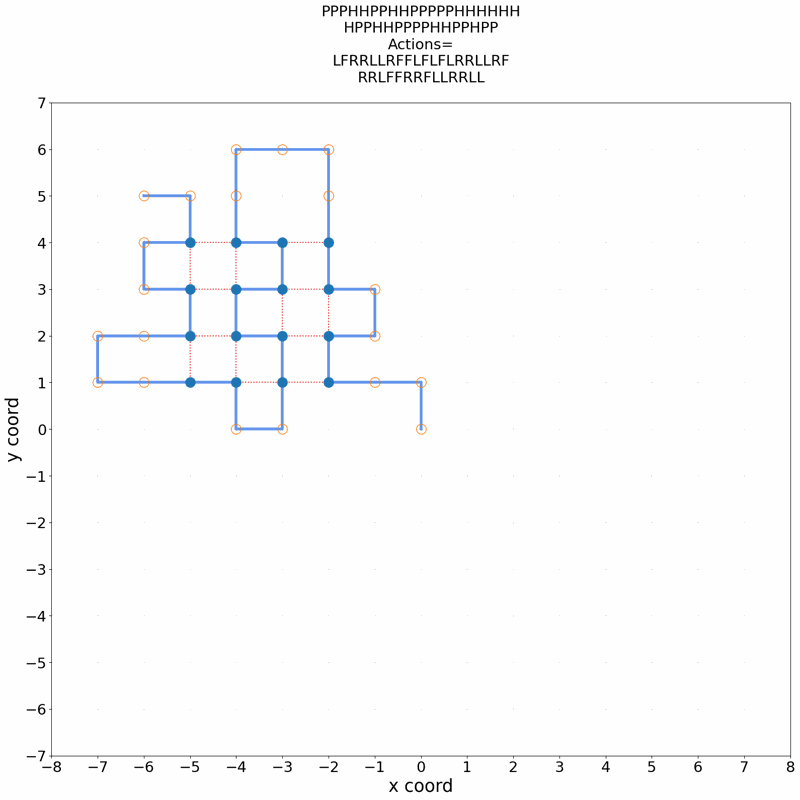
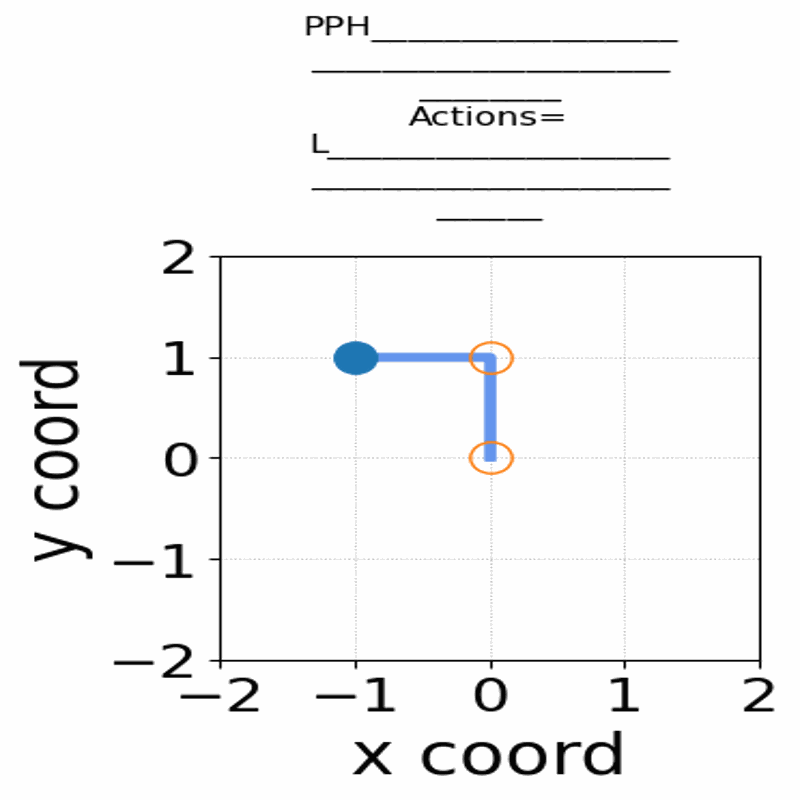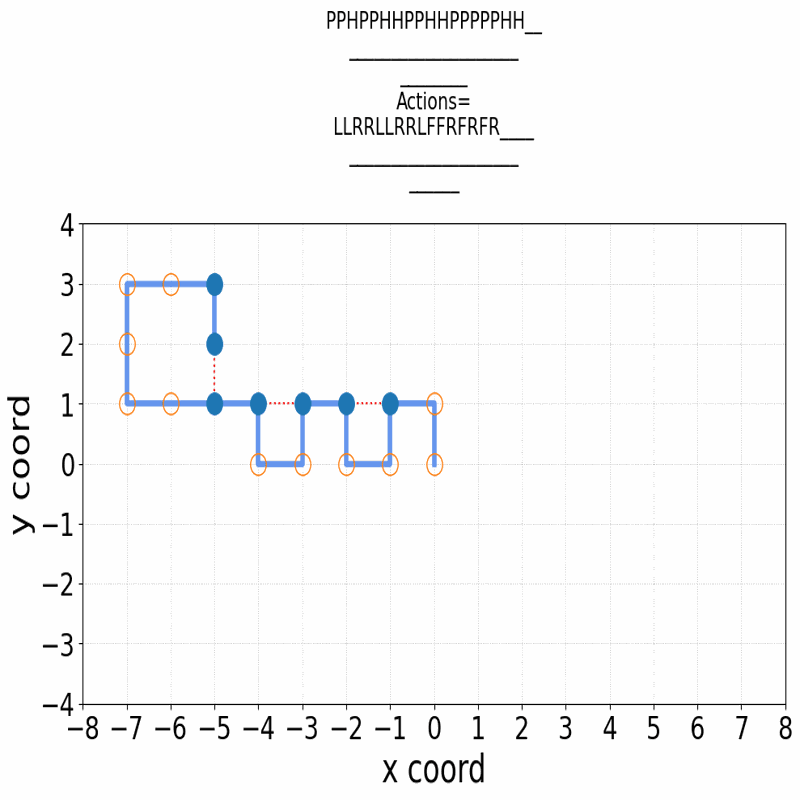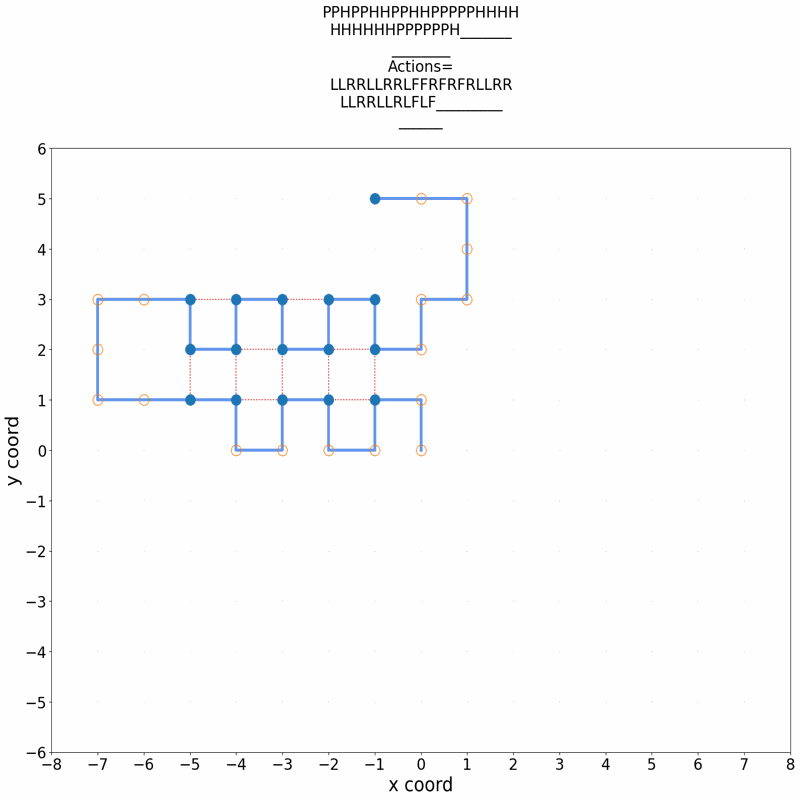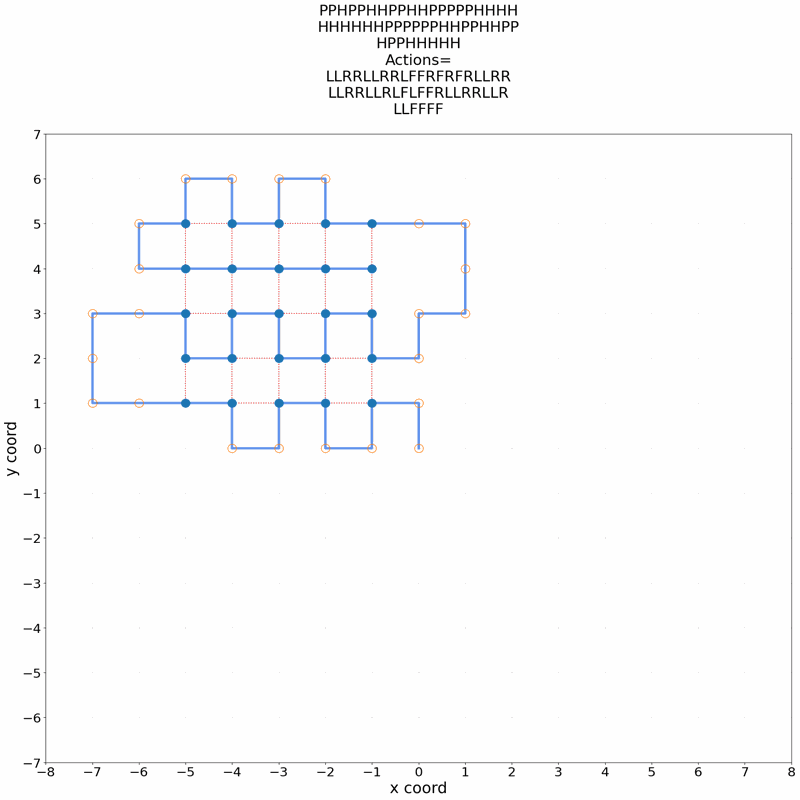
 Gif of 48mer folding to a conformation with best-known enegry of -23
Gif of 48mer folding to a conformation with best-known enegry of -23
This repository contains source code for the publication:
Applying Deep Reinforcement Learning to the HP Model for Protein Structure Prediction
- Extended abstract is accepted at the Machine Learning and the Physical Sciences workshop, NeurIPS 2022 (website)
NeurIPS Poster
https://neurips.cc/media/PosterPDFs/NeurIPS%202022/56887.png
NeurIPS Extended Abstract
https://ml4physicalsciences.github.io/2022/files/NeurIPS_ML4PS_2022_20.pdf
- Ful manuscript is published in journal Physica A: Statistical Mechanics and its Applications, Volume 609, 1 January 2023, 128395:
DOI of Physica A
Preprint on Arxiv:
@article{yang2022applying,
title = {Applying deep reinforcement learning to the HP model for protein structure prediction},
journal = {Physica A: Statistical Mechanics and its Applications},
volume = {609},
pages = {128395},
year = {2023},
issn = {0378-4371},
doi = {https://doi.org/10.1016/j.physa.2022.128395},
url = {https://www.sciencedirect.com/science/article/pii/S0378437122009530},
author = {Kaiyuan Yang and Houjing Huang and Olafs Vandans and Adithya Murali and Fujia Tian and Roland H.C. Yap and Liang Dai}
}showing the distinct best-known and next best conformations:
Zenodo open data repository
The conformation action sequences (in text files) are also available here in the folder ./conformations
conda v22.9 create specific city23 environment
conda create -n localCity23 python=3.8.10
# from conda env run the following
python --version
# Python 3.8.10
# install conda pytorch 1.10.1 cu11.3
conda install pytorch==1.10.1 torchvision==0.11.2 torchaudio==0.10.1 cudatoolkit=11.3 -c pytorch -c conda-forge
conda install -c conda-forge gym==0.21.0 numpy==1.21.4 matplotlib==3.5.0 scikit-learn==1.0.1 scipy==1.7.3 prettytable==2.4.0After installation, go to Waseda_Gym_Lattice/cwkx_DQN_lattice2d.py
cd Waseda_Gym_Lattice
# usage: cwkx_DQN_lattice2d.py [seq] [seed] [algo] [num_episodes] [use_early_stop]..
CUDA_VISIBLE_DEVICES=0 nohup python3 cwkx_DQN_lattice2d.py hhphphphphhhhphppphppphpppphppphppphphhhhphphphphh 42 50mer-DQN-Seed42-600K 600000 0 &- To inspect distinct local optimas, change this
# for local optima inspection
local_optima = {21, 23} # set of local optimas to inspect- To change greedy-epsilon decay mode:
# Exploration parameters
max_exploration_rate = 1
min_exploration_rate = 0.01
warmRestart = True
decay_mode = "exponential" # exponential, cosine, linear
num_restarts = 1 # for cosine decay warmRestart=True
exploration_decay_rate = 5 # for exponential decay
start_decay = 0 # for exponential and linear- To change NN arch
# choice of network for DRL = "FCN_QNet, RNN_LSTM_onlyLastHidden, BRNN..."
network_choice = "RNN_LSTM_onlyLastHidden"
# number of nodes in the hidden layers
hidden_size = 256
num_layers = 2If you need to change the FCN internal layers, go to Waseda_Gym_Lattice/minimalRL_DQN.py
Go to Waseda_Gym_Lattice/train_random_lattice2d.py
cd Waseda_Gym_Lattice
# usage: train_random_lattice2d.py [seq] [seed] [algo] [num_episodes] [use_early_stop]...
nohup python3 train_random_lattice2d.py hphpphhphpphphhpphph 42 20merA-RAND-Seed42-100K 100000 0 &An example inference script is provided for 20mer-B, 36mer, and 48mer at Waseda_Gym_Lattice/cwkx_DQN_inference.py
Select which N-mer to run the inference for by change N_mer:
N_mer = "48mer"
if N_mer == "20mer-B":
seq = "hhhpphphphpphphphpph" # 20mer-B
q_state_dict_path = "./Sample_DQN_weights/256L2/HHHPPH-PhyA_20merB-LSTM_noTrap_noES-1991-100K-seed1991-100000epi-state_dict.pth"
elif N_mer == "36mer":
seq = "PPPHHPPHHPPPPPHHHHHHHPPHHPPPPHHPPHPP" # 36mer
q_state_dict_path = "./Sample_DQN_weights/256L2/PPPHHP-36mer-LSTM_noTrap_noES-1991-500K-seed1991-500000epi-state_dict.pth"
elif N_mer == "48mer":
seq = "PPHPPHHPPHHPPPPPHHHHHHHHHHPPPPPPHHPPHHPPHPPHHHHH" # 48mer
# 2-layer LSTM architecture (in 256L2 dir)
q_state_dict_path = "./Sample_DQN_weights/256L2/48mer-256L2-example-state_dict.pth"
# 3-layer LSTM architecture (in 512L3 dir)
q_state_dict_path = "./Sample_DQN_weights/512L3/48mer-512L3-example-state_dict.pth"Essentailly, the training script cwkx_DQN_lattice2d.py saves the model as a state_dict.
cwkx_DQN_inference.py loads the model for inference.
Four example trained weights for can be loaded from the Waseda_Gym_Lattice/Sample_DQN_weights/ folder. Three example weights for 256L2 architecture (for 20mer-B, 36mer, or 48mer), and an example weight for 512L3 architecture (for 48mer).
After choosing the trained weight and N-mer, simply run the script by:
cd Waseda_Gym_Lattice
python cwkx_DQN_inference.pyRL Agent and Env adapted from Lester James V. Miranda's repo gym-lattice:
Visualization code has snippets from John S Schreck's repo RLFold:
DQN code adapted from
- PyTorch DQN Tutorial: https://pytorch.org/tutorials/intermediate/reinforcement_q_learning.html
- Chris G. Willcocks's lecture and tutorial: https://www.youtube.com/watch?v=BNSwFURmaCA&list=PLMsTLcO6ettgmyLVrcPvFLYi2Rs-R4JOE&index=3
- MinimalRL DQN: https://github.com/seungeunrho/minimalRL/blob/master/dqn.py
RNN code adapted from Aladdin Persson YouTube video: