v0.7.0 --- STILL IN ACTIVE DEV
30x `fastq` to SNV`vcf` at $3.34 EC2 costs, completes in 57m & process thousands of genomes an hour-
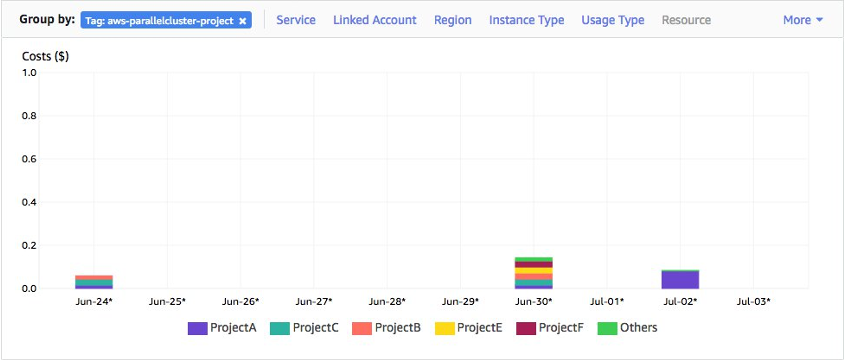
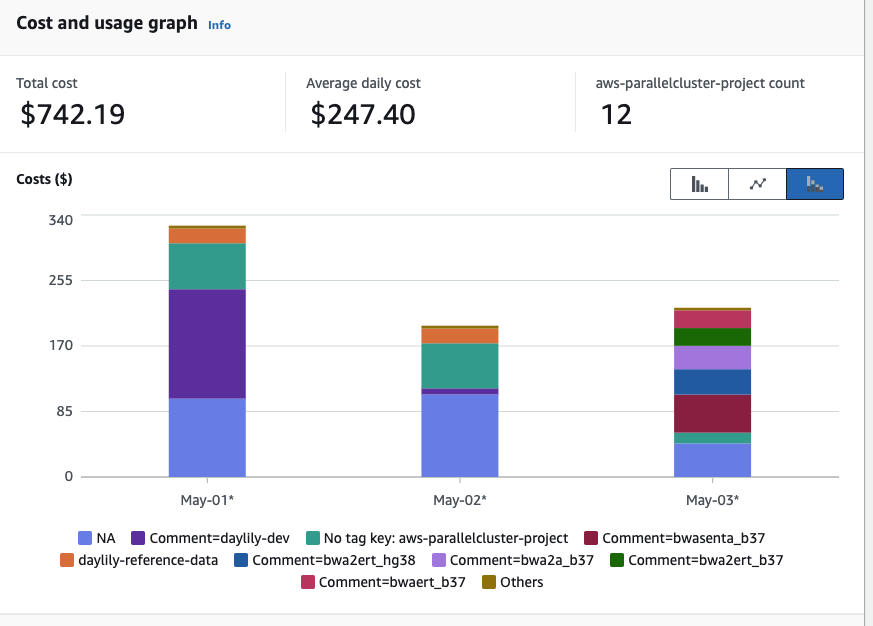
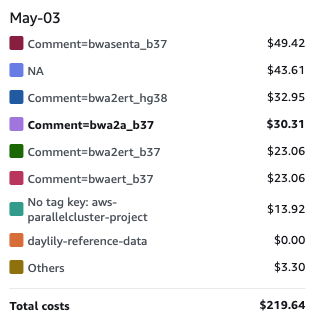
PLUS SNV/SV calling options at other sensitivities / extensive sample + batch QC reporting / performance & cost reporting + budgeting
-
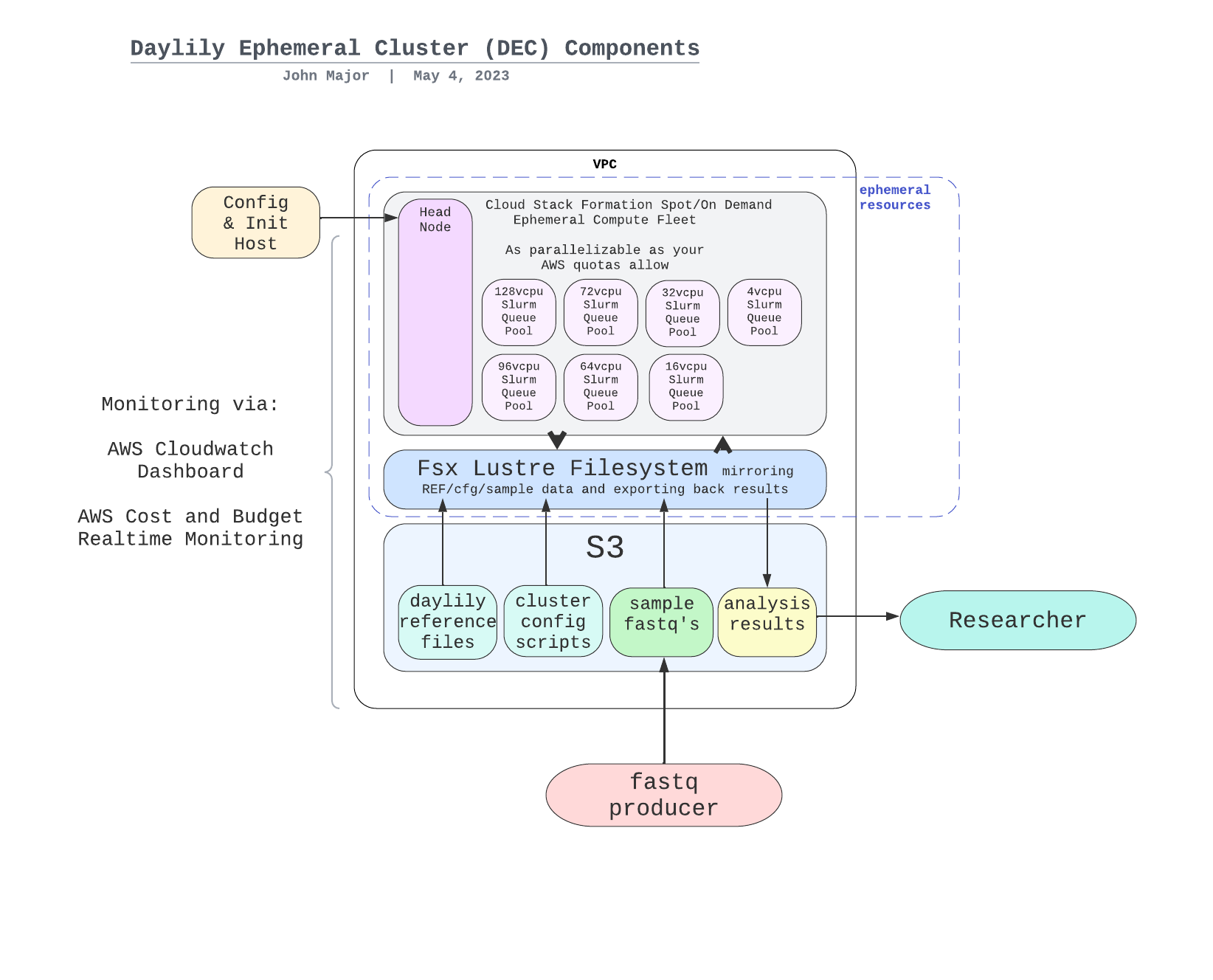
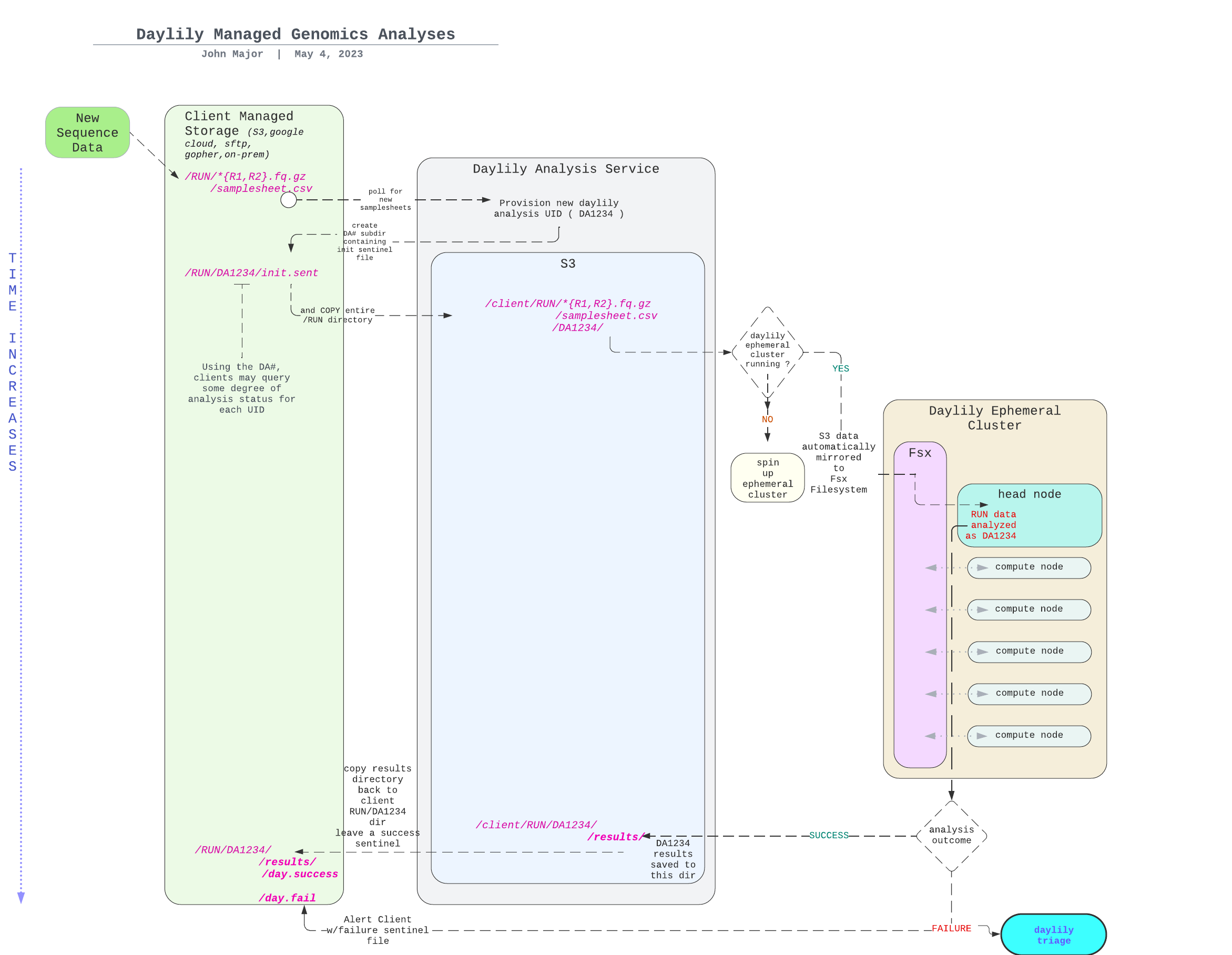
Daylily provides a single point of contact to the myriad systems which need to be orchestrated in order to run omic analysis reproducibly, reliably and at scale in the cloud. All you need is a laptop and access to an AWS console. After a FULL INSTALLATION you will be ready to begin processing up to thousands of genomes an hour (pending your AWS quotas).
-
Daylily is open source and free to use(excepting the Sentieon pipeline licensing fees which will be added to that pipeline). I hope some neat tricks I deploy are of help to others see blog.
Note Daylily Informatics is available for consulting services to integrate daylily into your operations, migrate pipelines into this framework, optimize existing pipelines, or general informatics work. daylily@daylilyinformatics.com
-
Daylily Informatics offers a managed genomic analysis service where, depending on the analyses and TAT desired, you pay a per-sample fee for daylily to run the desired analysis.
-
The gist of the standard deployment can be reviewed here.
-
Please contact daylily@daylilyinformatics.com for further information.
Before getting into the cool informatics business going on, there is a boatload of complex ops systems running to manage EC2 spot instances, navigate spot markets, as well as mechanisms to monitor and observe all aspects of this framework. AWS ParallelCluster is the glue holding everything together, and deserves special thanks.
The system is designed to be robust, secure, auditable, and should only take a matter of days to stand up. Please contact me for further details.
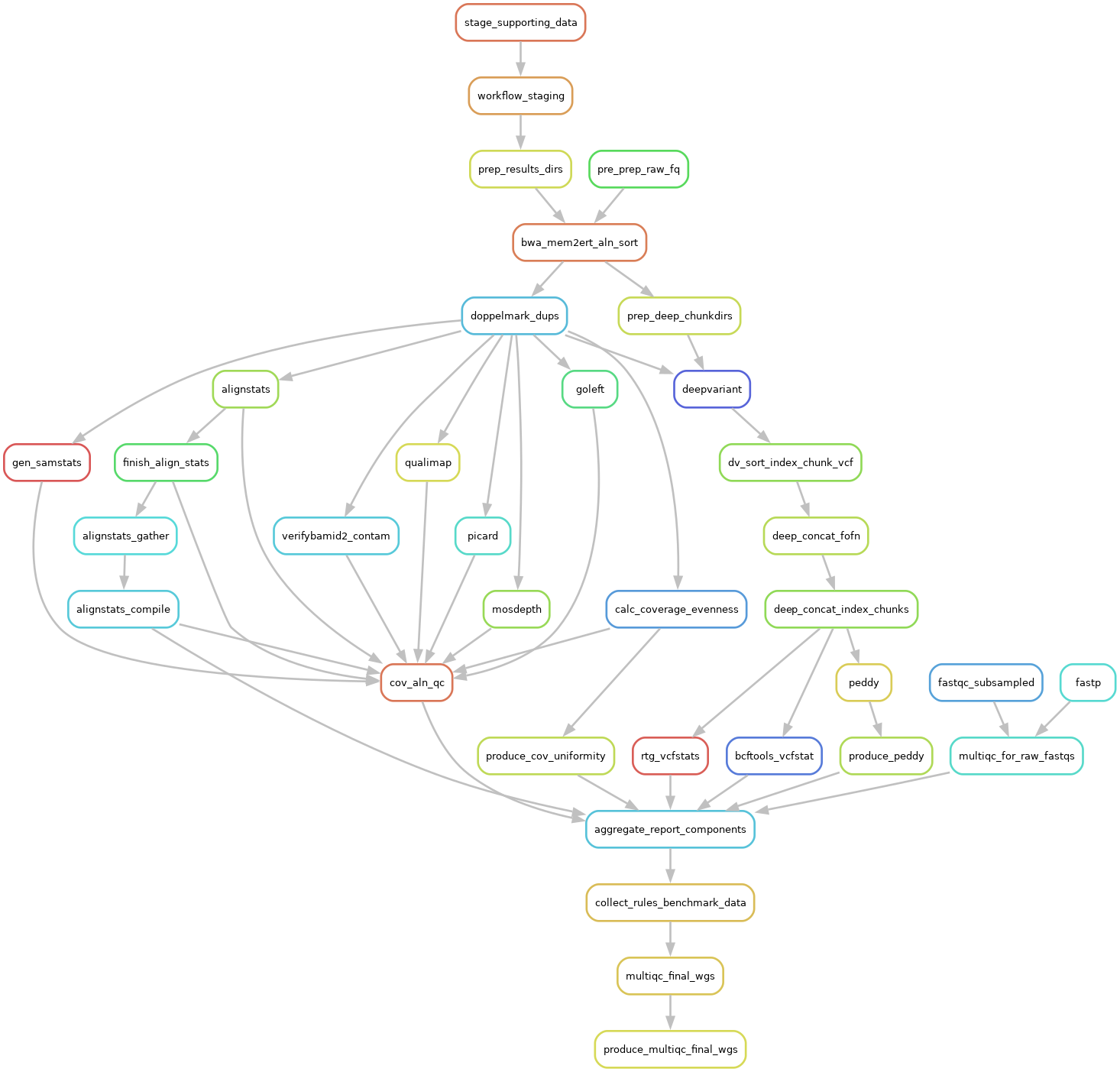
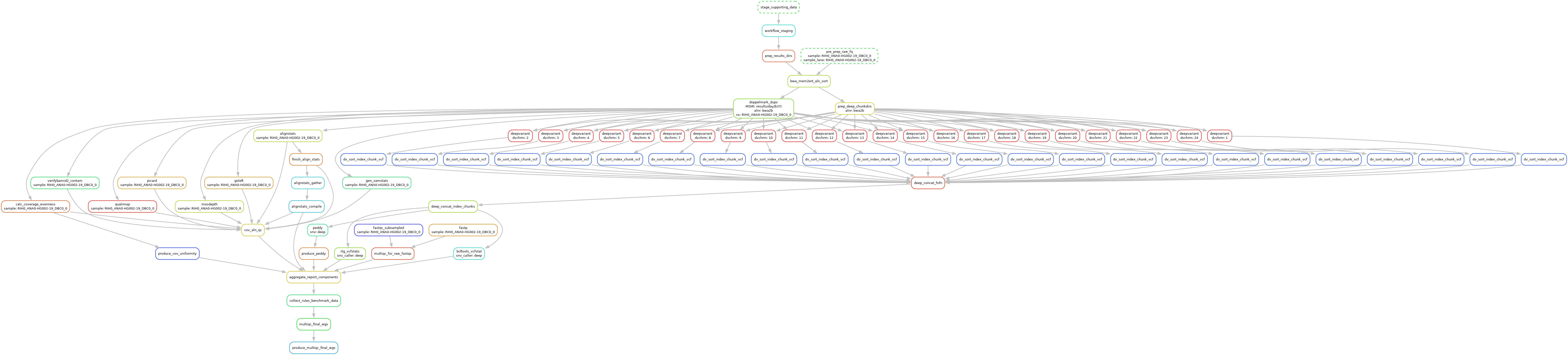
The DAG For 1 Sample Running Through The BWA-MEM2ert+Doppelmark+Deepvariant+Manta+TIDDIT+Dysgu+Svaba+QCforDays Pipeline
NOTE: each node in the below DAG is run as a self-contained job. Each job/node/rule is distributed to a suitable EC2 spot(or on demand if you prefer) instance to run. Each node is a packaged/containerized unit of work. This dag represents jobs running across sometimes thousands of instances at a time. Slurm and Snakemake manage all of the scaling, teardown, scheduling, recovery and general orchestration: cherry on top: killer observability & per project resource cost reporting and budget controls!
-
The above is actually a compressed view of the jobs managed for a sample moving through this pipeline. This view is of the dag which properly reflects parallelized jobs.
Daylily was built while drawing on over 20 years of experience in clinical genomics and informatics. These principles were kept front and center while building this framework.
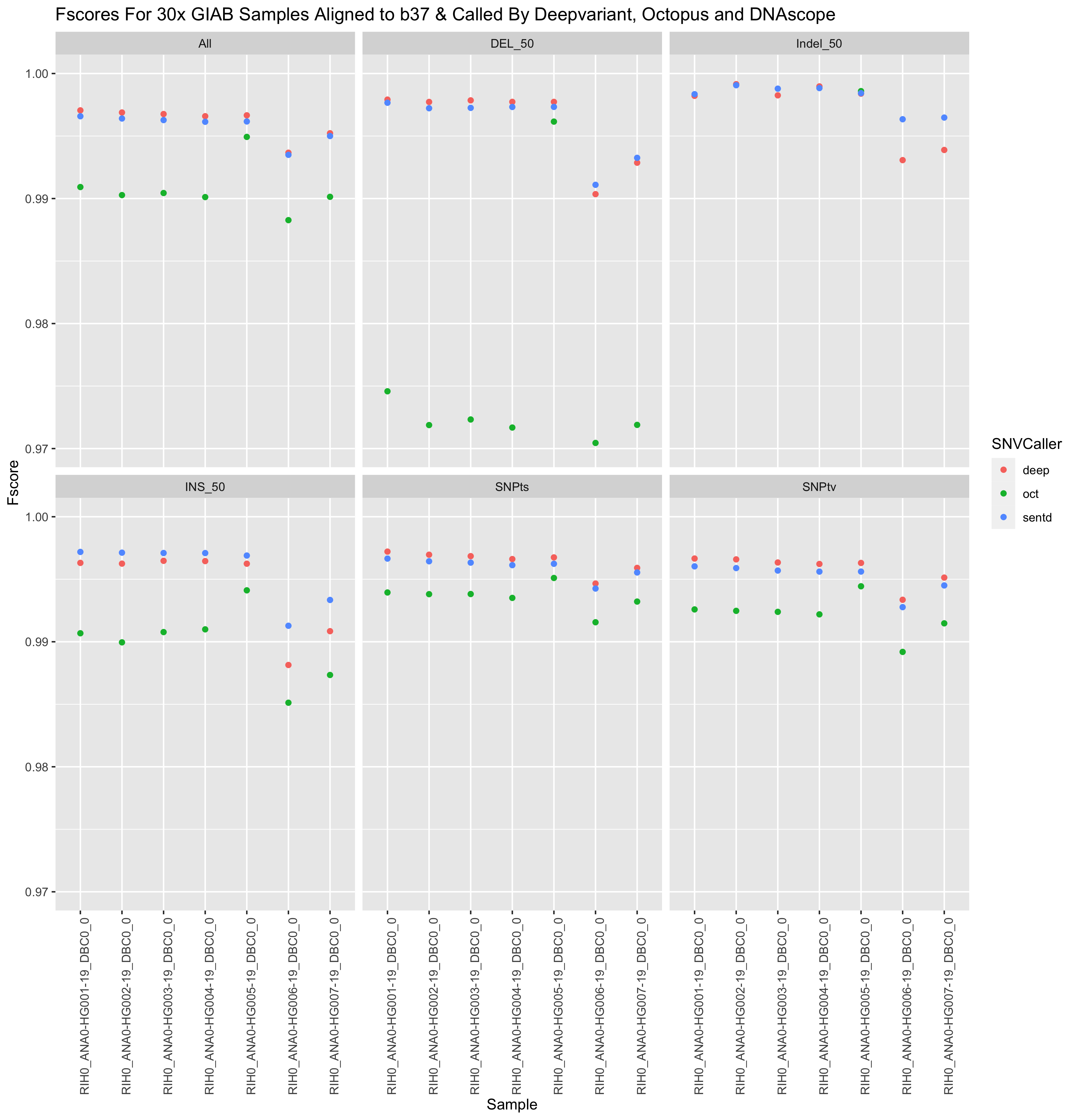
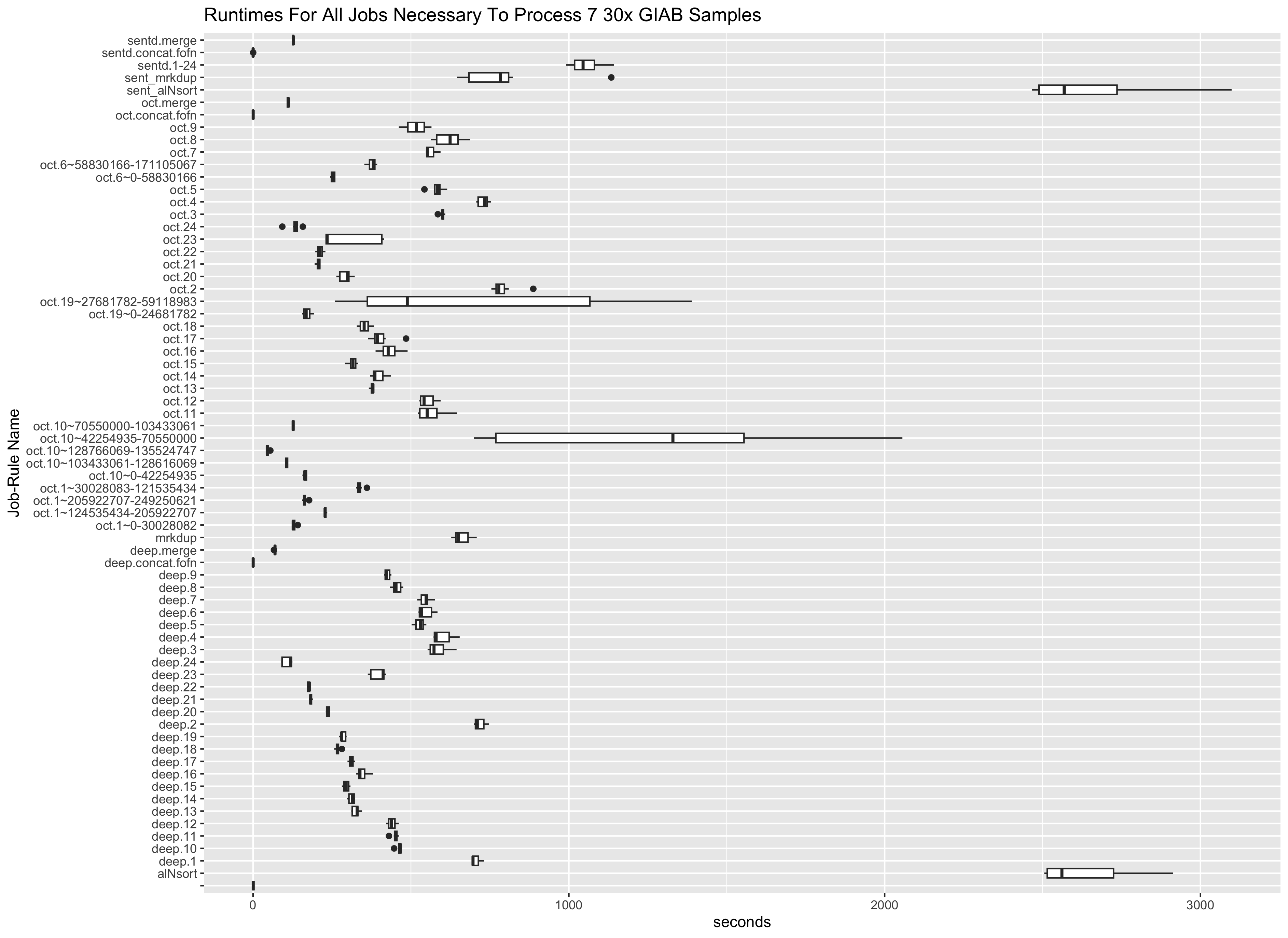
Presented below are Fscores, runtime and costs to run 3 pipelines. The results below are generated from the google-brain 30x Novaseq fastqs for all 7 GIAB samples. These fastqs and an analysis_manifest are included in the daylily-references S3 bucket so you may run these samples to show concordance with results shown here. The tools chosen for inclusion in daylily have been heavily optimized for speed and accuracy. The reported results are the median across all 7 GIAB samples. Costs are the average EC2 spot instance price to process fq.gz->snv.vcf per sample.
| Pipeline | SNPts/SNPtv fscore | INS fscore | DEL fscore | Indel fscore | e2e walltime | e2e instance min | Avg EC2 Cost |
|---|---|---|---|---|---|---|---|
| Sentieon** BWA + SentDeDup + DNAscope (BD) | 0.996 / 0.996 | 0.997* | 0.997 | 0.998* | 61m | 68m* | $3.34^*1 - 128vcpu |
| BWA-MEM2 + DpplDeDup + Octopus (B2O) | 0.994 / 0.992 | 0.991 | 0.971 | 0.800 | 72.4m | 273m | $12.92 - various vcpu |
| BWA-MEM2 + DpplDeDup + Deepvariant (B2D) | 0.997 / 0.996* | 0.996 | 0.998* | 0.998* | 57m* | 156m | $8.54 - 128 vcpu |
^=s/w licensing required to run the sentieon tool
*=highest value
The batch is comprised of google-brain Novaseq 30x HG002 fastqs, and again downsampling to: 25,20,15,10,5x.
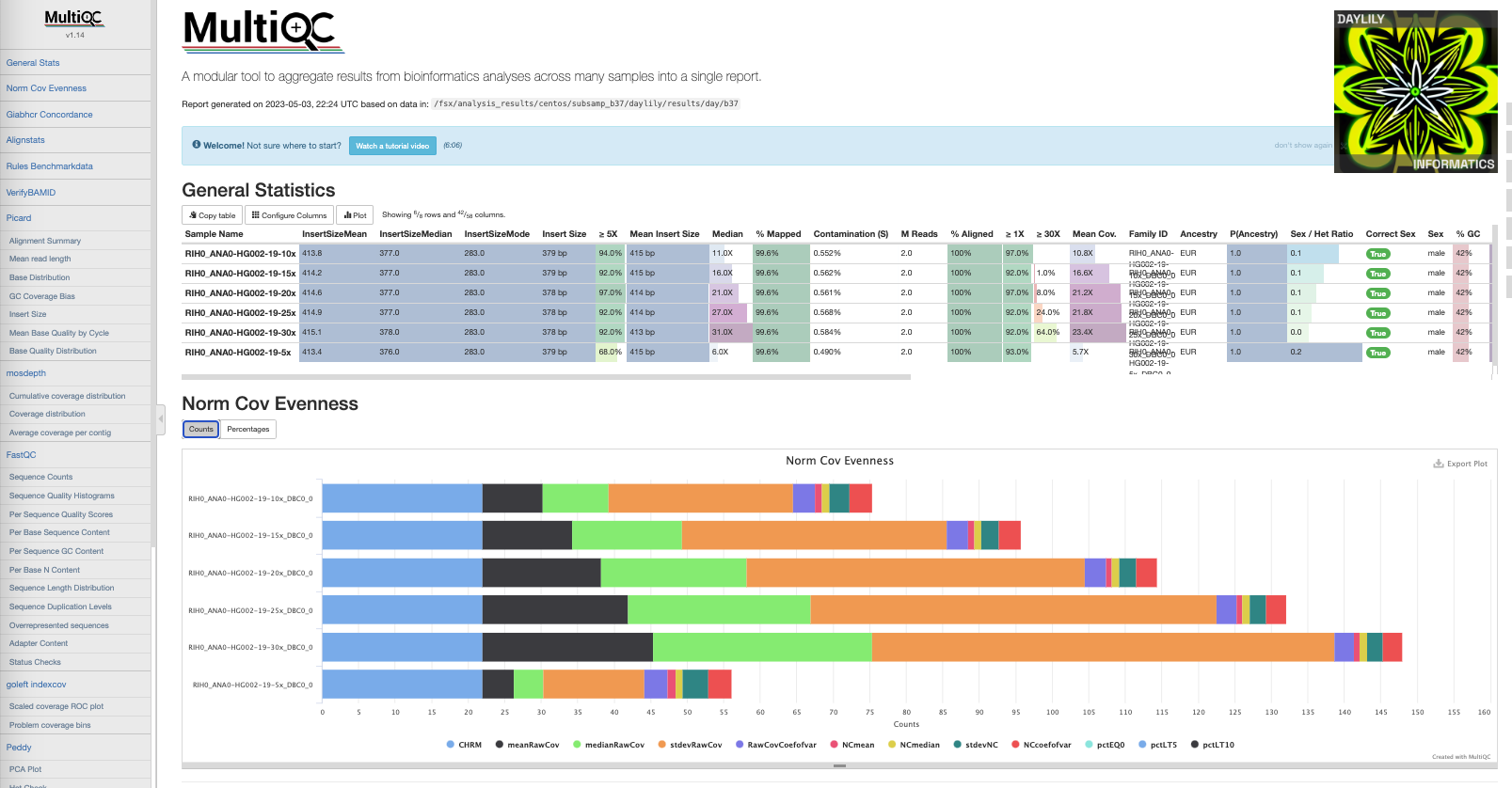
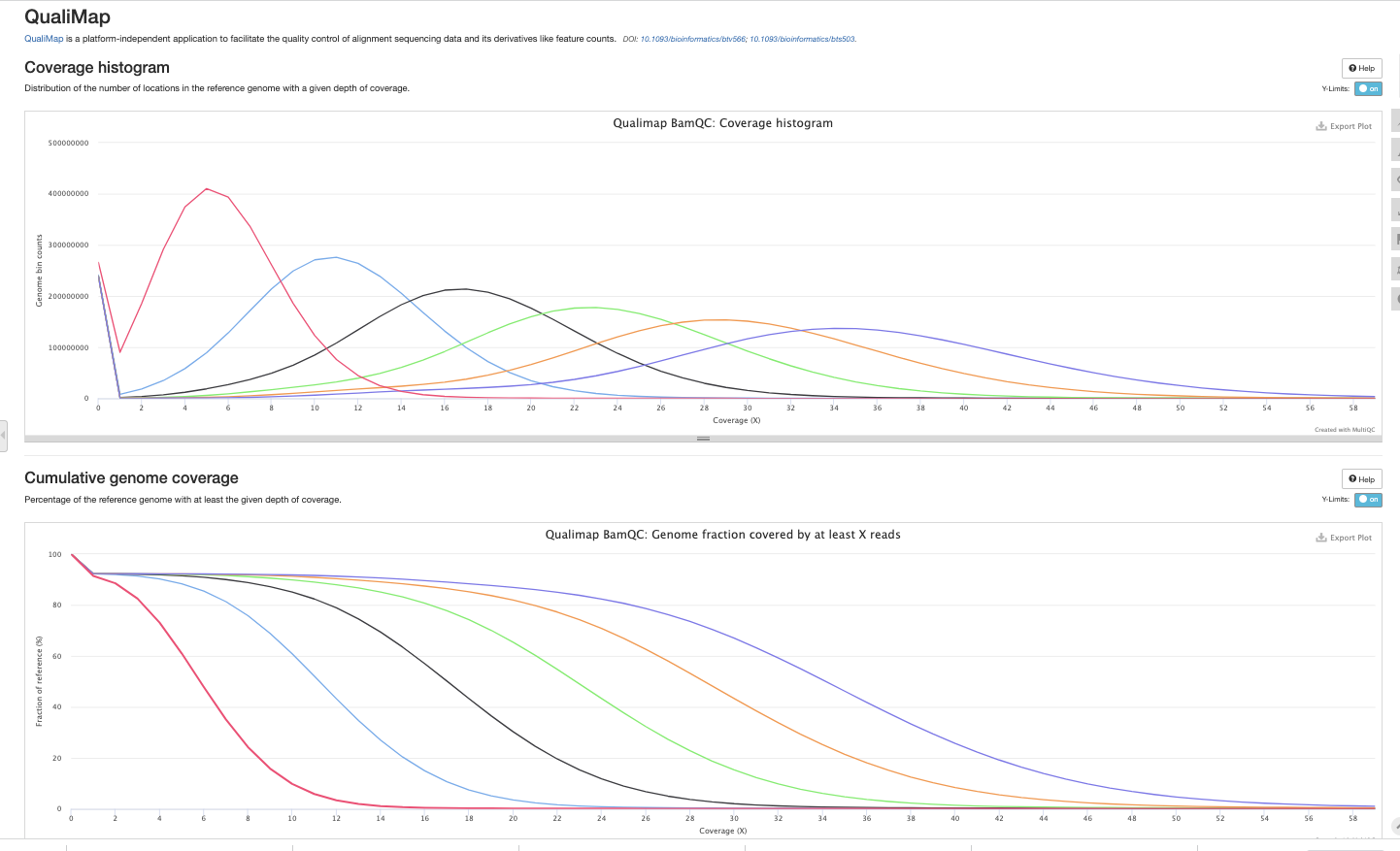
Example report.
- A visualization of just the directories (minus log dirs) created by daylily
b37 shown, hg38 is supported as well
- [with files](docs/ops/tree_full.md
Reported faceted by: SNPts, SNPtv, INS>0-<51, DEL>0-51, Indel>0-<51. Generated when the correct info is set in the analysis_manifest.
Picture and list of tools
- snakemake github action tests.
- Structural Variant Calling Concordance Analysis For The SV Callers:
- Manta
- TIDDIT
- Svaba
- Dysgu
- Octopus (which is a good small SV caller)
- Annotation of SNV / SV
vcffiles with potentially clinically relevant info (VEP is in testing). - Document the steps to quickly re-run the 7 30x GIAB samples from scratch.
- Explore hybrid assemblies using short and long reads (ONT + PacBio).
named in honor of Margaret Oakley Dahoff
1: plus Sentieon licensing fees
I'm getting cromwell running w/in the AWS ParallelCluster framework. This will allow for the running of WDLs in the cloud using the self-scaling cluster defined here. I am not keen on trying to get things to work similarly between snakemake and cromwell. For now these things are co-habitating, but I think the next cleanup steps will be to break this repo into three: the aws pcluster bits, snakemake stuff and cromwell.
For now, docs for cromwell will follow as soon as I have clean base cases running locally and in ParallelCluster.
Fail to copy still
2024/08/25 23:07:43 NOTICE: human_GRCh38_ens105/aligner_indices/star-fusion_1.10.1_index.zip: Skipped copy as --dry-run is set (size 32.394Gi) 2024/08/25 23:07:43 NOTICE: human_GRCh38_ens105/aligner_indices/star_2.7.8a_index.zip: Skipped copy as --dry-run is set (size 26.090Gi) 2024/08/25 23:07:43 NOTICE: human_GRCh38_ens105/aligner_indices/star_2.7.0f_index/SA: Skipped