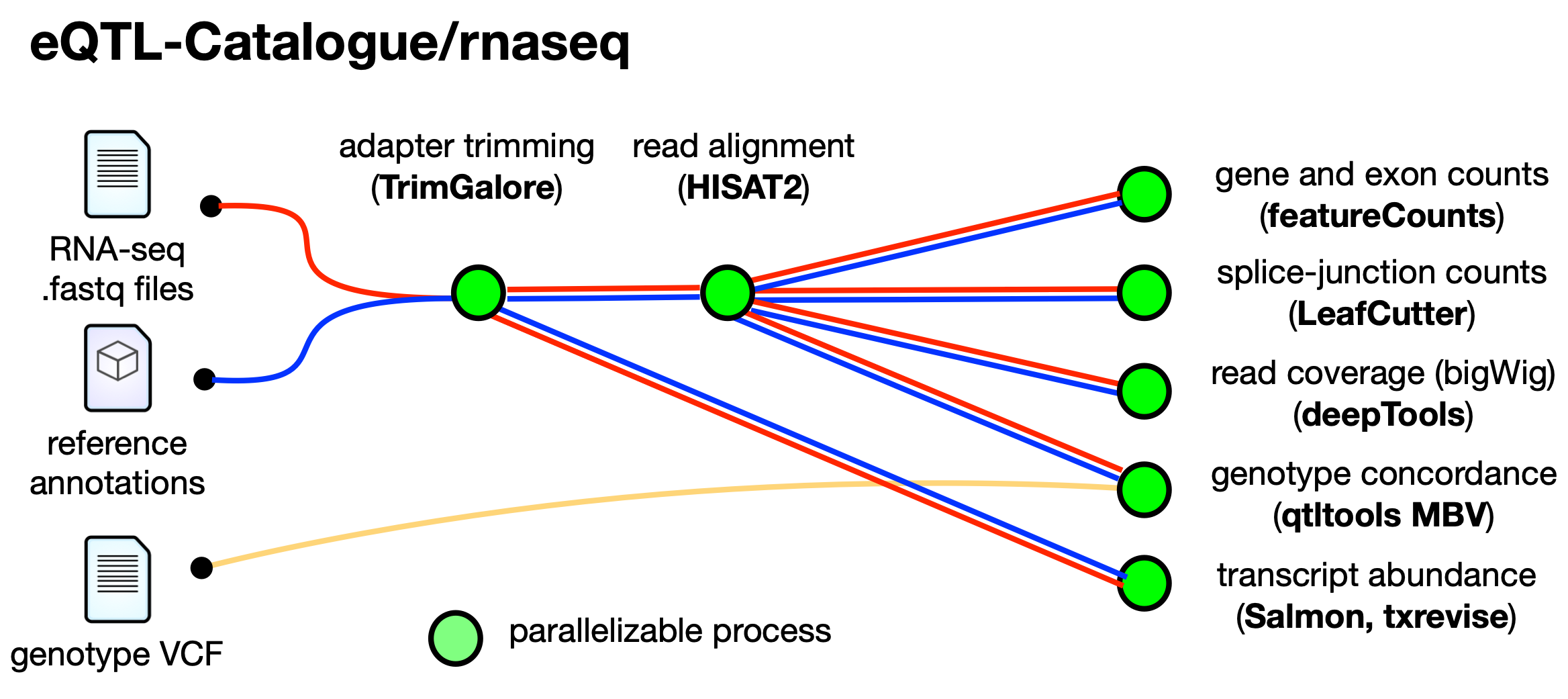
eQTL-Catalogue/rnaseq is a bioinformatics analysis pipeline used for processing RNA-sequencing data for the eQTL Catalogue.
The workflow processes raw data from fastq inputs (Trim Galore!); aligns the reads (HiSAT2); generates gene and exon counts (featureCounts, DEXSeq); quantifes transcript usage (Salmon), transcriptional event usage (txrevise) and splice junction usage (leafcutter); and check concordance between genotypes in BAM and VCF files (qtltools mbv).
The pipeline is built using Nextflow, a bioinformatics workflow tool to run tasks across multiple compute infrastructures in a very portable manner. It comes with docker / singularity containers making installation trivial and results highly reproducible.
The eQTL-Catalogue/rnaseq pipeline comes with documentation about the pipeline, found in the docs/ directory:
The schema shown below represents the high level structure of the pipeline.
This pipeline is highly influenced by much earlier version of the nf-core/rnaseq pipeline which was originally written for use at the National Genomics Infrastructure, part of SciLifeLab in Stockholm, Sweden, by Phil Ewels (@ewels) and Rickard Hammarén (@Hammarn).
New quantification methods (exon expression, transcript usage, transcriptional event usage and intron-splicing usage) are added by Alasoo Lab within the OpenTargets eQTL Catalogue project. Please cite eQTL Catalogue paper if this resource have been used for your research. https://doi.org/10.1038/s41588-021-00924-w
Many thanks to other who have helped out along the way too, including (but not limited to): @Galithil, @pditommaso, @orzechoj, @apeltzer, @colindaven.