Ultrafast prediction of somatic structural variations by filtering out reads matched to pan-genome k-mer sets
Efficient deTection of CHromosomal rearrangements and fusIoN Genes
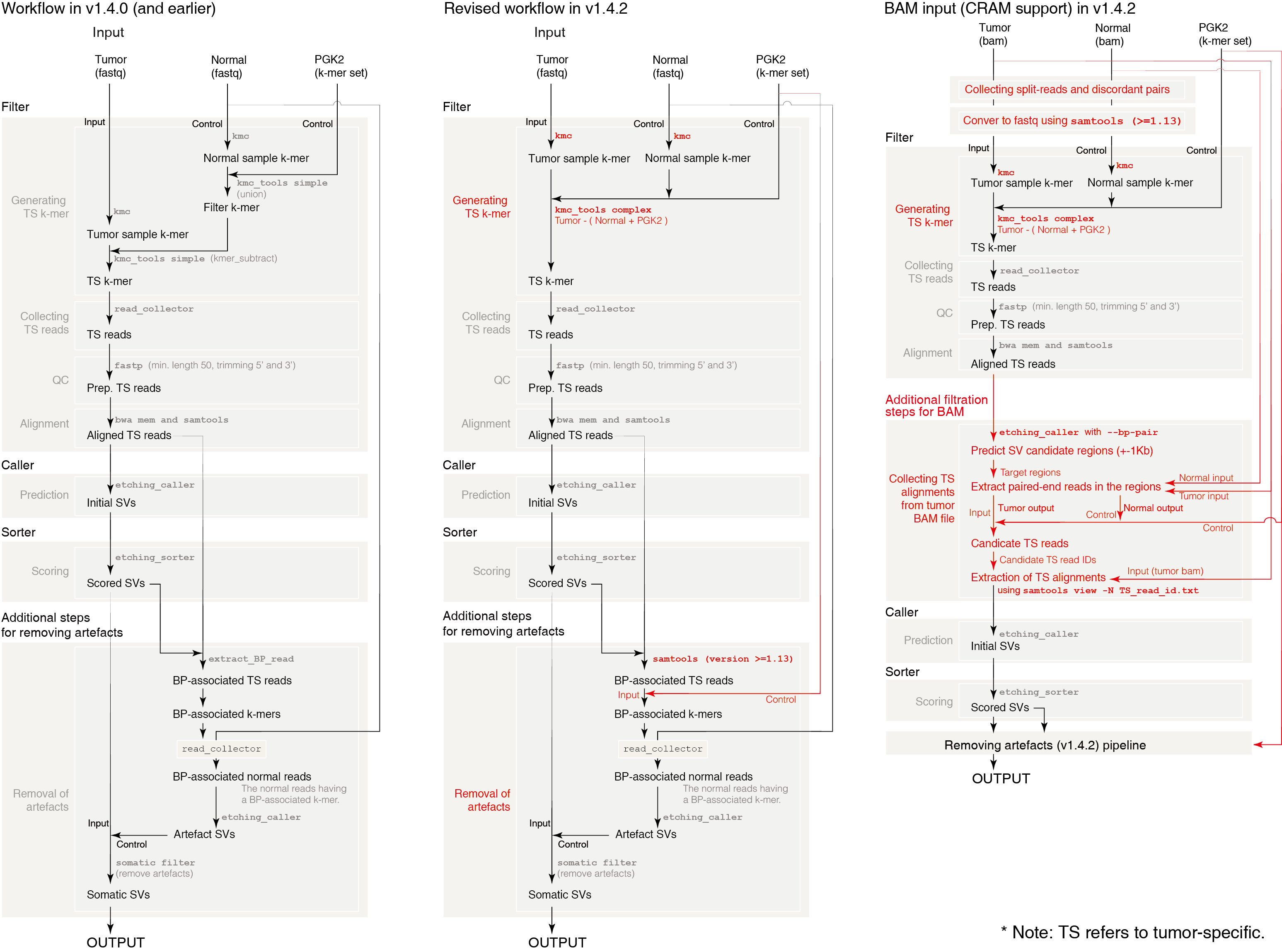
ETCHING takes about 3 hours for WGS data with 30X normal and 50X tumor on 30 threads on DELL 930 server. You can also find codes, k-mer set, and DEMO files on our website.
http://big.hanyang.ac.kr/ETCHING/download.html
We recommend using the new version of the pan-genome k-mer set, PGK2. You can download it from our website.
Using PGK2, ETCHING can detect somatic SVs from tumor sequencing data without matched-normal in comparable performance with other cutting-edge SV detection tools.
- Requirement changed
- g++ >=7 (>=7.3 recommended)
- cmake >=3.14
- samtools >=1.13 (using htslib >=1.13)
- BAM mode accelerated
- CRAM support
- Minor bugs fixed
See CHANGE.md for older updates.
ETCHING was a bit slow in removing remaining germline SVs after scoring SVs. We solved this issue by increasing Samtools requirement to >=1.13 for collecting the paired-end reads relevant to the remaining artifacts. As we updated the step, we accelerated the workflow for BAM input along with CRAM support.
- 64-bit LINUX with >=32GB RAM (at least >=16GB).
- Tested on Fedora workstation, Centos, and Ubuntu
-
Required to compile
-
gcc, g++ (>=7), make, cmake (>=3.14), wget
Note: g++ >=7.3 recommended
-
-
Required to run
- BWA, samtools (>=1.13) using htslib (>=1.13)
We prepared a simple guide for CentOS/Fedora or Ubuntu/Debian/Mint users. You can skip this step if all requirements were installed.
Note: We tested this guide on Fedora32/33/34, CentOS7/8, Ubuntu16.04/18.04/20.04/22.04, Mint19/20, Debian11, and MX Linux.
# Required programs
sudo yum install -y gcc gcc-c++ make cmake bwa samtools wget## Required programs
sudo apt install -y gcc g++ make cmake bwa samtools wgetOnce the requirements were solved, you can install ETCHING as follows.
# Download ETCHING
git clone --depth=1 https://github.com/ETCHING-team/ETCHING.git
# Move to /path/to/ETCHING
cd ETCHING
# Installation (g++ >=7 and cmake >=3.14 required)
# g++ >=7.3 recommended
make
# Set your environment for ETCHING
echo "export PATH=$PATH:/path/to/ETCHING/bin" >> ~/.bashrc
exec $SHELL# Change directory
cd /wherever/you/want/
# Download and decompress DEMO
wget http://big.hanyang.ac.kr/ETCHING/tiny_demo.tar.gz
# or you can find a demo file from our website
# http://big.hanyang.ac.kr/ETCHING/
# Decompress
tar zxvf tiny_demo.tar.gz
cd tiny_demo
# Run demo
etching -1 tumor_1.fq -2 tumor_2.fq -1c normal_1.fq -2c normal_2.fq -g Chr22.fa -f demo_PGKYou can run with bam or cram.
# or
etching -b tumor.sorted.cram -bc normal.sorted.cram -g Chr22.fa -f demo_PGK -p cram_mode_outputThe Pan-Genome k-mer(PGK) set is used to build PGK filter. Since we updated the PGK to PGK2, you can also download the PGK2 as below. If you have no matched-normal data, you are highly recommended to use the PGK2 instead of PGK to call somatic SVs.
# Move to etching directory
mkdir -p /path/to
cd /path/to
# Download
wget http://big.hanyang.ac.kr/ETCHING/PGK2.tar.gz
# or you can download it from Amazon S3
# wget https://biglabhyu.s3.amazonaws.com/ETCHING/PGK.tar.gz
# Decompress
tar zxvf PGK2.tar.gz
cd PGK2
# Then, you must see PGK2.kmc_pre and PGK2_kmc_suf
ls /path/to/PGK2/*
/path/to/PGK2/PGK2.kmc_pre
/path/to/PGK2/PGK2.kmc_sufTo use PGK2 (recommended), add -f /path/to/PGK2/PGK2 option.
For instance,
etching -1 tumor_1.fq -2 tumor_2.fq -g reference.fa -f /path/to/PGK2/PGK2Then, etching script will find /path/to/PGK2/PGK2.kmc_pre and /path/to/PGK2/PGK2.kmc_suf.
Alternatively, you can make your custom k-mer set using make_pgk included in ETCHING package.
In the absence of matched-normal sample, we recommend using >=200 genomes to build your custom k-mer set excluding rare k-mers (<1% allele frequency).
If you have 600 genomes, use -m 6 option, which is the minimum frequency of k-mer to be included in your k-mer set.
make_pgk -i 600_genome.list -m 6 -o my_pgkHere, 600_genome.list is a file of FASTA files of genomes in the format of
/path/to/references/hg19.fasta
/path/to/references/GRCh38.fa
/path/to/references/NA12878.chr1.fna
/path/to/references/NA12878.chr2.fna
.
.
.
/path/to/assemblies/someones_genome.scaffold_1.fa
/path/to/assemblies/someones_genome.scaffold_2.fa
docker
# Download ETCHING docker image
wget http://big.hanyang.ac.kr/ETCHING/ETCHING_v1.4.2.docker.tar
# Load the image
docker load -i ETCHING_v1.4.2.docker.tar
# Check the image
docker imagesYou can see like below (numbers can be different)
| REPOSITORY | TAG | IMAGE ID | CREATED | SIZE |
|---|---|---|---|---|
| etching | 1.4.2 | 967b3d7fb6f7 | 40 hours ago | 1.5GB |
Download demo data and run ETCHING with docker
wget http://big.hanyang.ac.kr/ETCHING/tiny_demo.tar.gz
tar zxvf tiny_demo.tar.gz
docker run -i -t --rm -v $PWD/tiny_demo/:/work/ etching:1.4.2 etching \
-1 tumor_1.fq -2 tumor_2.fq -1c normal_1.fq -2c normal_2.fq -g Chr22.fa -f demo_PGK -t 8Here, etching:1.4.2 is REPOSITORY:TAG of ETCHING docker image.
To use your data, replace $PWD/tiny_demo with /your/data/path/. The directory also should contain k-mer set (PGK2).
Note: Keep /work/ in the above command line, since it's the default working directory.
An older version of ETCHING (v1.4.0) is also available on Amazon Web Service (AMI ID: ami-07c7a7d8934784df9; Region: us-east-1 (Northern Virginia)).
# Lunch EC2 instance from ETCHING AMI
# And connect to EC2 instance
ssh -i Your_Key ubuntu@Your_Instance_Address
# Decompress
tar zxvf ~/resources/DEMO.tar.gz
# Run demo
cd ~/resources/DEMO
bash example.shhttps://github.com/ETCHING-team/LR_Filter
https://github.com/ETCHING-team/etching_bench
Sohn, Ji., Choi, MH., Yi, D. et al. Ultrafast prediction of somatic structural variations by filtering out reads matched to pan-genome k-mer sets. Nat. Biomed. Eng (2022). https://doi.org/10.1038/s41551-022-00980-5
Jang-il Sohn, Min-Hak Choi, Dohun Yi, A. Vipin Menon, and Jin-Wu Nam
Bioinformatics and Genomics Lab., Dept. of Life Science, Hanyang University, Seoul 04763, Korea
Jin-Wu Nam (jwnam@hanyang.ac.kr)
Jang-il Sohn (sohnjangil@gmail.com)
Min-Hak Choi (choiminhak1004@gmail.com)
Dohun Yi (kutarballoon@gmail.com)