This package pools together information from different databases and APIs in order to annotate SNPs and genes. In particular this uses databases by:
To install:
devtools::install_github("KatrionaGoldmann/omicAnnotations")
First, if you want to include associated diseases from disGeNET you will need to get an api_key. To get this sign up and get your API key either from the API directly or run:
api_key <- get_api_key(email="your email", password="your password")
For example for the entire gene summary:
gene_df <- gene_summary(genes=c("MS4A1", "FMOD", "FGF1", "SLAMF6"),
diseases="C20",
disease_api_token=api_key)
## [1] "Annotating from self-curated data..."
## [1] "Getting gene summaries..."
## [1] "Finding associated diseases..."
gene_df <- gene_df[, c("Gene", "description", "summary", "Associated_diseases")]
kable(gene_df, format = "markdown", row.names = FALSE)
| Gene | description | summary | Associated_diseases |
|---|---|---|---|
| MS4A1 | membrane spanning 4-domains A1 | This gene encodes a member of the membrane-spanning 4A gene family. Members of this nascent protein family are characterized by common structural features and similar intron/exon splice boundaries and display unique expression patterns among hematopoietic cells and nonlymphoid tissues. This gene encodes a B-lymphocyte surface molecule which plays a role in the development and differentiation of B-cells into plasma cells. This family member is localized to 11q12, among a cluster of family members. Alternative splicing of this gene results in two transcript variants which encode the same protein. [provided by RefSeq, Jul 2008] | Common Variable Immunodeficiency; Acquired Hypogammaglobulinemia; Immunoglobulin Deficiency, Late-Onset |
| FMOD | fibromodulin | Fibromodulin belongs to the family of small interstitial proteoglycans. The encoded protein possesses a central region containing leucine-rich repeats with 4 keratan sulfate chains, flanked by terminal domains containing disulphide bonds. Owing to the interaction with type I and type II collagen fibrils and in vitro inhibition of fibrillogenesis, the encoded protein may play a role in the assembly of extracellular matrix. It may also regulate TGF-beta activities by sequestering TGF-beta into the extracellular matrix. Sequence variations in this gene may be associated with the pathogenesis of high myopia. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jun 2013] | |
| FGF1 | fibroblast growth factor 1 | The protein encoded by this gene is a member of the fibroblast growth factor (FGF) family. FGF family members possess broad mitogenic and cell survival activities, and are involved in a variety of biological processes, including embryonic development, cell growth, morphogenesis, tissue repair, tumor growth and invasion. This protein functions as a modifier of endothelial cell migration and proliferation, as well as an angiogenic factor. It acts as a mitogen for a variety of mesoderm- and neuroectoderm-derived cells in vitro, thus is thought to be involved in organogenesis. Multiple alternatively spliced variants encoding different isoforms have been described. [provided by RefSeq, Jan 2009] | |
| SLAMF6 | SLAM family member 6 | The protein encoded by this gene is a type I transmembrane protein, belonging to the CD2 subfamily of the immunoglobulin superfamily. This encoded protein is expressed on Natural killer (NK), T, and B lymphocytes. It undergoes tyrosine phosphorylation and associates with the Src homology 2 domain-containing protein (SH2D1A) as well as with SH2 domain-containing phosphatases (SHPs). It functions as a coreceptor in the process of NK cell activation. It can also mediate inhibitory signals in NK cells from X-linked lymphoproliferative patients. Alternative splicing results in multiple transcript variants encoding distinct isoforms.[provided by RefSeq, May 2010] |
You can check for publications focusing on genes with given terms.
Either using associated_publications:
gene_pubs <- associated_publications(genes=c("FGF1"),
keywords=c("rheumatoid"),
split="OR",
verbose=TRUE)
kable(gene_pubs, format = "markdown", row.names=FALSE)
| Gene | Publications |
|---|---|
| FGF1 | The transcriptomic profiling of SARS-CoV-2 compared to SARS, MERS, EBOV, and H1N1.; sICAM-1 as potential additional parameter in the discrimination of the Sjögren syndrome and non-autoimmune sicca syndrome: a pilot study.; [Effects of Huatan Tongluo Recipe on IL-1β-induced Proliferation of Rheumatoid Arthritis Synovial Fibroblasts and the Production of TNF-α and aFGF].; Fibroblast growth factors, fibroblast growth factor receptors, diseases, and drugs.; VEGF, FGF1, FGF2 and EGF gene polymorphisms and psoriatic arthritis.; Transcription factor Ets-1 regulates fibroblast growth factor-1-mediated angiogenesis in vivo: role of Ets-1 in the regulation of the PI3K/AKT/MMP-1 pathway.; Induction of RANKL expression and osteoclast maturation by the binding of fibroblast growth factor 2 to heparan sulfate proteoglycan on rheumatoid synovial fibroblasts.; Acidic fibroblast growth factor in synovial cells.; Characterization of tissue outgrowth developed in vitro in patients with rheumatoid arthritis: involvement of T cells in the development of tissue outgrowth.; Fibroblast growth factor-1 (FGF-1) enhances IL-2 production and nuclear translocation of NF-kappaB in FGF receptor-bearing Jurkat T cells.; A novel in vitro assay for human angiogenesis.; Expression and functional expansion of fibroblast growth factor receptor T cells in rheumatoid synovium and peripheral blood of patients with rheumatoid arthritis.; Detection of T cells responsive to a vascular growth factor in rheumatoid arthritis.; Coexpression of phosphotyrosine-containing proteins, platelet-derived growth factor-B, and fibroblast growth factor-1 in situ in synovial tissues of patients with rheumatoid arthritis and Lewis rats with adjuvant or streptococcal cell wall arthritis.; Platelet-derived growth factors and heparin-binding (fibroblast) growth factors in the synovial tissue pathology of rheumatoid arthritis.; Fibroblast growth factors: from genes to clinical applications.; Production of platelet derived growth factor B chain (PDGF-B/c-sis) mRNA and immunoreactive PDGF B-like polypeptide by rheumatoid synovium: coexpression with heparin binding acidic fibroblast growth factor-1.; Detection of high levels of heparin binding growth factor-1 (acidic fibroblast growth factor) in inflammatory arthritic joints. |
Or gene_summary:
gene_df <- gene_summary(genes=c("FGF1"),
associated_diseases =FALSE,
gene_description=FALSE,
publications = TRUE)
## [1] "Annotating from self-curated data..."
## [1] "Getting publications from PubMed..."
kable(gene_df, format = "markdown", row.names=FALSE)
| Gene | Type | Curated_description | Publications |
|---|---|---|---|
| FGF1 | Fibroblast Growth Factors | FGF/FGFR Pathways in Multiple Sclerosis and in Its Disease Models.; The transcriptomic profiling of SARS-CoV-2 compared to SARS, MERS, EBOV, and H1N1.; sICAM-1 as potential additional parameter in the discrimination of the Sjögren syndrome and non-autoimmune sicca syndrome: a pilot study.; Oligodendroglial fibroblast growth factor receptor 1 gene targeting protects mice from experimental autoimmune encephalomyelitis through ERK/AKT phosphorylation.; [Effects of Huatan Tongluo Recipe on IL-1β-induced Proliferation of Rheumatoid Arthritis Synovial Fibroblasts and the Production of TNF-α and aFGF].; Dysregulation of pathways involved in the processing of cancer and microenvironment information in MCA + TPA transformed C3H/10T1/2 cells.; Fibroblast growth factors, fibroblast growth factor receptors, diseases, and drugs.; VEGF, FGF1, FGF2 and EGF gene polymorphisms and psoriatic arthritis.; Cutaneous gene expression by DNA microarray in murine sclerodermatous graft-versus-host disease, a model for human scleroderma.; Transcription factor Ets-1 regulates fibroblast growth factor-1-mediated angiogenesis in vivo: role of Ets-1 in the regulation of the PI3K/AKT/MMP-1 pathway.; Angiocidal effect of Cyclosporin A: a new therapeutic approach for pathogenic angiogenesis.; Induction of RANKL expression and osteoclast maturation by the binding of fibroblast growth factor 2 to heparan sulfate proteoglycan on rheumatoid synovial fibroblasts.; Acidic fibroblast growth factor in synovial cells.; Characterization of tissue outgrowth developed in vitro in patients with rheumatoid arthritis: involvement of T cells in the development of tissue outgrowth.; Lack of FGF-1 overexpression during autoimmune nephritis in the kidneys of MRL lpr/lpr mice.; Fibroblast growth factor-1 (FGF-1) enhances IL-2 production and nuclear translocation of NF-kappaB in FGF receptor-bearing Jurkat T cells.; Cloning and characterization of a novel upstream untranslated exon of the mouse Fgf-1 gene.; Cloning and characterization of the mouse Fgf-1 gene.; A novel in vitro assay for human angiogenesis.; Expression and functional expansion of fibroblast growth factor receptor T cells in rheumatoid synovium and peripheral blood of patients with rheumatoid arthritis.; Environmental influences on fatty acid composition of membranes from autoimmune MRL lpr/lpr mice.; Costimulation of human CD4+ T cells by fibroblast growth factor-1 (acidic fibroblast growth factor).; Detection of T cells responsive to a vascular growth factor in rheumatoid arthritis.; Coexpression of phosphotyrosine-containing proteins, platelet-derived growth factor-B, and fibroblast growth factor-1 in situ in synovial tissues of patients with rheumatoid arthritis and Lewis rats with adjuvant or streptococcal cell wall arthritis.; Platelet-derived growth factors and heparin-binding (fibroblast) growth factors in the synovial tissue pathology of rheumatoid arthritis.; Fibroblast growth factors: from genes to clinical applications.; Production of platelet derived growth factor B chain (PDGF-B/c-sis) mRNA and immunoreactive PDGF B-like polypeptide by rheumatoid synovium: coexpression with heparin binding acidic fibroblast growth factor-1.; Detection of high levels of heparin binding growth factor-1 (acidic fibroblast growth factor) in inflammatory arthritic joints. |
Looks for enriched pathways with gene sets using enrichR.
lymphoid_pathways <- enriched_pathways(
genes=c("LAMP5", "LINC01480", "FAM92B", "SLAMF6", "CEP128",
"FKBP11", "CRTAM", "ISG20", "ZBP1", "TMEM229B",
"FAM46C", "XBP1", "APOBEC3G", "TNIK", "CD2", "SP140",
"ACOXL", "PTPRCAP", "PDCD1", "KCNN3", "GZMK",
"IGFLR1", "SH2D2A", "PIM2", "TPST2"),
libraries = c('Pathways'),
dbs=NULL,
check_for_updates = FALSE)
If that doesn’t work it may be because the website is down. This happens occasionally. You can check by using:
listEnrichrDbs()
Plots
lymphoid_pathways$plot
eqtl_table <- associated_eqtl(genes=c("ENSG00000164308"), p_cutoff=1)
## [1] "Looking at SNPs"
## [1] "Looking at Genes"
kable(eqtl_table, row.names=F)
| rsid | chromosome | molecular\_trait\_id | gene\_id | tissue | qtl\_group | pvalue | neg\_log10\_pvalue | se | beta | median\_tpm | study\_id | type | alt | position | ac | maf | variant | ref | r2 | an |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs57584041 | 5 | ENSG00000164308 | ENSG00000164308 | CL\_0000235 | macrophage\_IFNg | 0.142464 | 0.8462949 | 0.862435 | 1.2793100 | 14.576 | Alasoo\_2018 | SNP | C | 95877044 | 5 | 0.0297619 | chr5\_95877044\_T\_C | T | 0.87667 | 168 |
| rs6556892 | 5 | ENSG00000164308 | ENSG00000164308 | CL\_0000235 | macrophage\_IFNg | 0.299611 | 0.5234422 | 0.338419 | 0.3536530 | 14.576 | Alasoo\_2018 | SNP | A | 95878071 | 56 | 0.3333330 | chr5\_95878071\_C\_A | C | 0.92606 | 168 |
| rs55763081 | 5 | ENSG00000164308 | ENSG00000164308 | CL\_0000235 | macrophage\_IFNg | 0.324924 | 0.4882182 | 1.103560 | -1.0940300 | 14.576 | Alasoo\_2018 | SNP | G | 95876702 | 3 | 0.0178571 | chr5\_95876702\_A\_G | A | 0.81827 | 168 |
| rs61540882 | 5 | ENSG00000164308 | ENSG00000164308 | CL\_0000235 | macrophage\_IFNg | 0.325028 | 0.4880792 | 1.103460 | -1.0936900 | 14.576 | Alasoo\_2018 | INDEL | T | 95876577 | 3 | 0.0178571 | chr5\_95876577\_TAAA\_T | TAAA | 0.81754 | 168 |
| rs796285486 | 5 | ENSG00000164308 | ENSG00000164308 | CL\_0000235 | macrophage\_IFNg | 0.325028 | 0.4880792 | 1.103460 | -1.0936900 | 14.576 | Alasoo\_2018 | INDEL | T | 95876577 | 3 | 0.0178571 | chr5\_95876577\_TAAA\_T | TAAA | 0.81754 | 168 |
| rs154457 | 5 | ENSG00000164308 | ENSG00000164308 | CL\_0000235 | macrophage\_IFNg | 0.347034 | 0.4596280 | 0.376044 | 0.3560110 | 14.576 | Alasoo\_2018 | SNP | A | 95876181 | 130 | 0.2261900 | chr5\_95876181\_G\_A | G | 0.93729 | 168 |
| rs154458 | 5 | ENSG00000164308 | ENSG00000164308 | CL\_0000235 | macrophage\_IFNg | 0.354738 | 0.4500923 | 0.375816 | 0.3501150 | 14.576 | Alasoo\_2018 | SNP | T | 95876288 | 130 | 0.2261900 | chr5\_95876288\_C\_T | C | 0.94067 | 168 |
| rs154456 | 5 | ENSG00000164308 | ENSG00000164308 | CL\_0000235 | macrophage\_IFNg | 0.355252 | 0.4494635 | 0.375702 | 0.3496330 | 14.576 | Alasoo\_2018 | SNP | T | 95876161 | 130 | 0.2261900 | chr5\_95876161\_A\_T | A | 0.94121 | 168 |
| rs144088066 | 5 | ENSG00000164308 | ENSG00000164308 | CL\_0000235 | macrophage\_IFNg | 0.358421 | 0.4456066 | 1.143510 | -1.0571400 | 14.576 | Alasoo\_2018 | INDEL | C | 95878406 | 3 | 0.0178571 | chr5\_95878406\_CTCT\_C | CTCT | 0.74292 | 168 |
| rs17085223 | 5 | ENSG00000164308 | ENSG00000164308 | CL\_0000235 | macrophage\_IFNg | 0.363755 | 0.4391910 | 1.139710 | -1.0419100 | 14.576 | Alasoo\_2018 | SNP | T | 95877713 | 3 | 0.0178571 | chr5\_95877713\_G\_T | G | 0.77062 | 168 |
| rs113842599 | 5 | ENSG00000164308 | ENSG00000164308 | CL\_0000235 | macrophage\_IFNg | 0.402339 | 0.3954079 | 1.153880 | -0.9722280 | 14.576 | Alasoo\_2018 | SNP | G | 95878112 | 3 | 0.0178571 | chr5\_95878112\_C\_G | C | 0.70249 | 168 |
| rs749046156 | 5 | ENSG00000164308 | ENSG00000164308 | CL\_0000235 | macrophage\_IFNg | 0.409355 | 0.3878999 | 0.958131 | 0.7952580 | 14.576 | Alasoo\_2018 | INDEL | GT | 95877403 | 5 | 0.0297619 | chr5\_95877403\_G\_GT | G | 0.65265 | 168 |
| rs397957177 | 5 | ENSG00000164308 | ENSG00000164308 | CL\_0000235 | macrophage\_IFNg | 0.409355 | 0.3878999 | 0.958131 | 0.7952580 | 14.576 | Alasoo\_2018 | INDEL | GT | 95877403 | 5 | 0.0297619 | chr5\_95877403\_G\_GT | G | 0.65265 | 168 |
| rs1256088833 | 5 | ENSG00000164308 | ENSG00000164308 | CL\_0000235 | macrophage\_IFNg | 0.482832 | 0.3162040 | 1.163830 | -0.8210970 | 14.576 | Alasoo\_2018 | INDEL | G | 95877403 | 2 | 0.0119048 | chr5\_95877403\_GT\_G | GT | 0.58191 | 168 |
| rs111471052 | 5 | ENSG00000164308 | ENSG00000164308 | CL\_0000235 | macrophage\_IFNg | 0.482832 | 0.3162040 | 1.163830 | -0.8210970 | 14.576 | Alasoo\_2018 | INDEL | G | 95877403 | 2 | 0.0119048 | chr5\_95877403\_GT\_G | GT | 0.58191 | 168 |
| rs154454 | 5 | ENSG00000164308 | ENSG00000164308 | CL\_0000235 | macrophage\_IFNg | 0.537536 | 0.2695924 | 0.385196 | 0.2386650 | 14.576 | Alasoo\_2018 | SNP | G | 95875943 | 133 | 0.2083330 | chr5\_95875943\_C\_G | C | 0.96320 | 168 |
| rs11372327 | 5 | ENSG00000164308 | ENSG00000164308 | CL\_0000235 | macrophage\_IFNg | 0.710863 | 0.1482141 | 0.818859 | -0.3047850 | 14.576 | Alasoo\_2018 | INDEL | AC | 95878741 | 162 | 0.0357143 | chr5\_95878741\_A\_AC | A | 0.91136 | 168 |
| rs397998782 | 5 | ENSG00000164308 | ENSG00000164308 | CL\_0000235 | macrophage\_IFNg | 0.710863 | 0.1482141 | 0.818859 | -0.3047850 | 14.576 | Alasoo\_2018 | INDEL | AC | 95878741 | 162 | 0.0357143 | chr5\_95878741\_A\_AC | A | 0.91136 | 168 |
| rs154459 | 5 | ENSG00000164308 | ENSG00000164308 | CL\_0000235 | macrophage\_IFNg | 0.738887 | 0.1314220 | 0.332555 | -0.1112910 | 14.576 | Alasoo\_2018 | SNP | T | 95876578 | 72 | 0.4285710 | chr5\_95876578\_A\_T | A | 0.94702 | 168 |
| rs154455 | 5 | ENSG00000164308 | ENSG00000164308 | CL\_0000235 | macrophage\_IFNg | 0.803819 | 0.0948417 | 0.335028 | -0.0835397 | 14.576 | Alasoo\_2018 | SNP | T | 95876057 | 74 | 0.4404760 | chr5\_95876057\_C\_T | C | 0.96802 | 168 |
omicAnnotations can also be used to find out more info about SNPs.
gwas_traits <- associated_traits(snps = c("rs2910686", "rs7329174"))
kable(gwas_traits, row.names = F)
| SNPs | Associated\_traits |
|---|---|
| rs2910686 | neutrophil count; ankylosing spondylitis; crohn’s disease; psoriasis; sclerosing cholangitis; ulcerative colitis |
| rs7329174 | systemic lupus erythematosus; crohn’s disease |
eqtl_table <- associated_eqtl(snps = c("rs2910686", "rs7329174"),
p_cutoff = 0.05)
## [1] "Looking at SNPs"
## [1] "Looking at Genes"
kable(eqtl_table, row.names = F)
| rsid | chromosome | molecular\_trait\_id | gene\_id | tissue | qtl\_group | pvalue | neg\_log10\_pvalue | se | beta | median\_tpm | study\_id | type | alt | position | ac | maf | variant | ref | r2 | an |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs2910686 | 5 | ENSG00000164308 | ENSG00000164308 | CL\_0000235 | macrophage\_IFNg | 0.0000000 | 32.033736 | 0.1084150 | 2.3491000 | 14.576 | Alasoo\_2018 | SNP | C | 96916885 | 68 | 0.4047620 | rs2910686 | T | 0.99787 | 168 |
| rs2910686 | 5 | ENSG00000164308 | ENSG00000164308 | CL\_0000235 | macrophage\_IFNg | 0.0000000 | 28.094117 | 0.1188220 | 2.2120400 | 14.576 | Alasoo\_2018 | SNP | C | 96916885 | 68 | 0.4047620 | rs2910686 | T | 0.99787 | 168 |
| rs2910686 | 5 | ENSG00000164307 | ENSG00000164307 | CL\_0000235 | macrophage\_IFNg | 0.0001333 | 3.875124 | 0.0474250 | -0.1917860 | 50.151 | Alasoo\_2018 | SNP | C | 96916885 | 68 | 0.4047620 | rs2910686 | T | 0.99787 | 168 |
| rs7329174 | 13 | ENSG00000102760 | ENSG00000102760 | UBERON\_0001013 | adipose\_naive | 0.0006357 | 3.196736 | 0.0714243 | 0.2470190 | 445.066 | FUSION | SNP | G | 40983974 | 38 | 0.0701107 | rs7329174 | A | 1.00000 | 542 |
| rs2910686 | 5 | ENSG00000247121 | ENSG00000247121 | CL\_0000235 | macrophage\_IFNg | 0.0007469 | 3.126725 | 0.0779628 | -0.2749620 | 1.260 | Alasoo\_2018 | SNP | C | 96916885 | 68 | 0.4047620 | rs2910686 | T | 0.99787 | 168 |
| rs2910686 | 5 | ENSG00000164307 | ENSG00000164307 | CL\_0000235 | macrophage\_IFNg | 0.0008048 | 3.094338 | 0.0611752 | -0.2143270 | 50.151 | Alasoo\_2018 | SNP | C | 96916885 | 68 | 0.4047620 | rs2910686 | T | 0.99787 | 168 |
| rs2910686 | 5 | ENSG00000113441 | ENSG00000113441 | CL\_0000235 | macrophage\_IFNg | 0.0012984 | 2.886608 | 0.0291856 | 0.0978188 | 14.738 | Alasoo\_2018 | SNP | C | 96916885 | 68 | 0.4047620 | rs2910686 | T | 0.99787 | 168 |
| rs7329174 | 13 | ENSG00000278390 | ENSG00000278390 | UBERON\_0009834 | brain | 0.0053281 | 2.273430 | 0.0627828 | -0.1757720 | 3.801 | BrainSeq | SNP | G | 40983974 | 39 | 0.0407098 | rs7329174 | A | 0.93510 | 958 |
| rs7329174 | 13 | ENSG00000102743 | ENSG00000102743 | UBERON\_0001013 | adipose\_naive | 0.0291754 | 1.534983 | 0.0500392 | -0.1097560 | 3.886 | FUSION | SNP | G | 40983974 | 38 | 0.0701107 | rs7329174 | A | 1.00000 | 542 |
| rs2910686 | 5 | ENSG00000113441 | ENSG00000113441 | CL\_0000235 | macrophage\_naive | 0.0482241 | 1.316736 | 0.0258004 | 0.0518735 | 14.738 | Alasoo\_2018 | SNP | C | 96916885 | 68 | 0.4047620 | rs2910686 | T | 0.99787 | 168 |
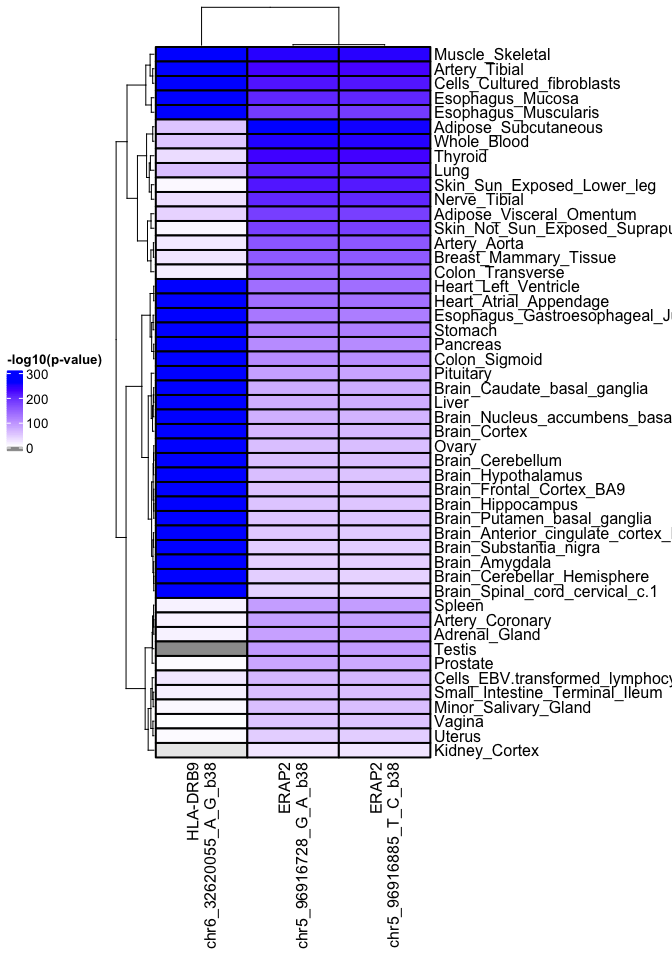
g2s <- data.frame("Genes"=c("ERAP2", "ERAP2", "HLA-DRB9"),
"Snps"=c("chr5_96916728_G_A", "chr5_96916885_T_C",
"chr6_32620055_A_G"))
df <- gtex_eqtl(gene_snp_pairs = g2s)
library(ComplexHeatmap)
hm <- gtex_heatmap(df)
draw(hm, heatmap_legend_side = "left")