PyViPR is a Jupyter widget that generates network dynamic and static visualizations of PySB, Tellurium, BNGL, SBML, and Ecell4 models using Cytoscape.js. Additionally, it can be used to visualize networks encoded in the graphml, sif, sbgn xml, cytoscape json, gexf, gml and yaml formats.
To try out PyViPR interactively in your web browser, just click on the binder link below:
> conda install pyvipr -c ortegas> conda install pyvipr -c ortegas
> jupyter labextension install @jupyter-widgets/jupyterlab-manager
> jupyter labextension install pyvipr> pip install pyvipr> pip install pyvipr
> jupyter labextension install @jupyter-widgets/jupyterlab-manager
> jupyter labextension install pyvipr$ git clone https://github.com/LoLab-VU/pyvipr.git
$ cd pyvipr
$ pip install .After installing the widget, it can be used by importing it in the Jupyter notebook. The widget is simple to use with PySB models, SimulationResult objects, Tellurium models and BNGL & SBML files.
PyViPR has three main interfaces: Graph formats, PySB, and a Tellurium.
This interface leverages NetworkX and Cytoscape.js to generate network visualizations of graphs encoded in different file formats.
import pyvipr.network_viz as nviz
nviz.graphml_view('path_to_file/mygraph.graphml', layout_name='fcose')PySB is needed to visualize PySB models and it is needed if you want to use the pyvipr.pysb_viz module:
Installing PySB from pip:
> pip install pysbWhen using pip the installation of PySB requires to manually install BioNetGen into the default path for your platform (/usr/local/share/BioNetGen on Mac and Linux, c:\Program Files\BioNetGen on Windows), or set the BNGPATH environment variable to the BioNetGen path on your machine.
Installing PySB from conda:
> conda install pysb -c alubbockimport pyvipr.pysb_viz as pviz
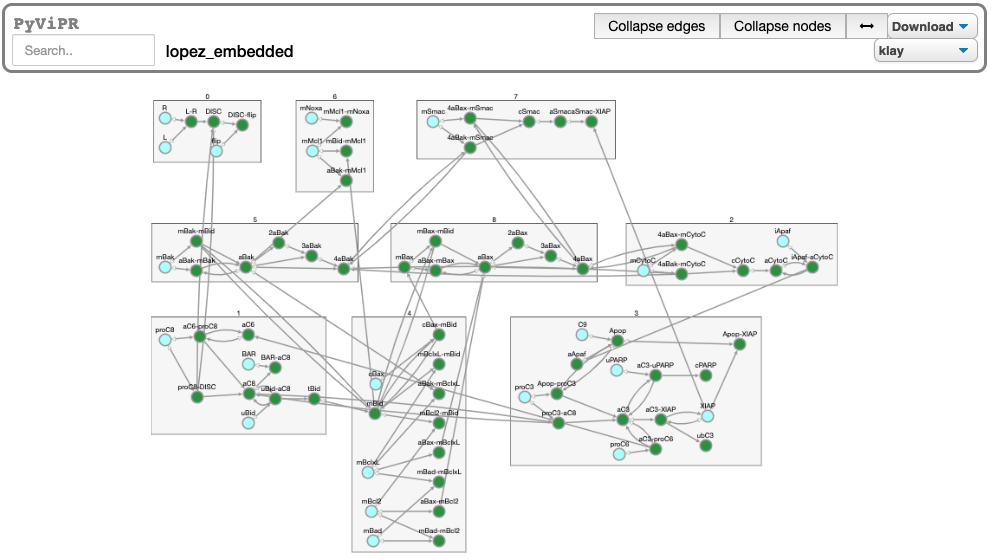
from pyvipr.examples_models.lopez_embedded import model
pviz.sp_comm_louvain_view(model, random_state=1, layout_name='klay')import pyvipr.pysb_viz as pviz
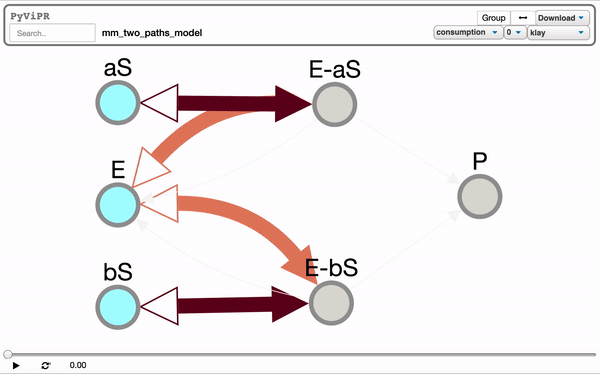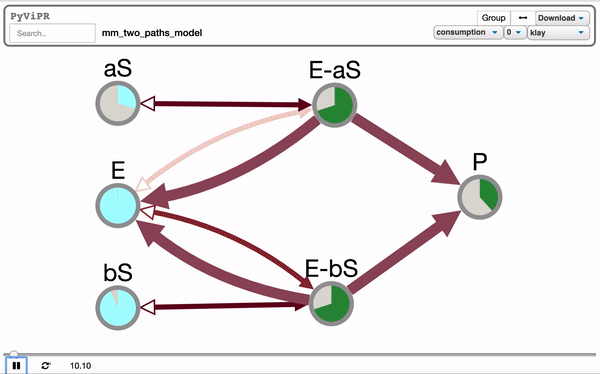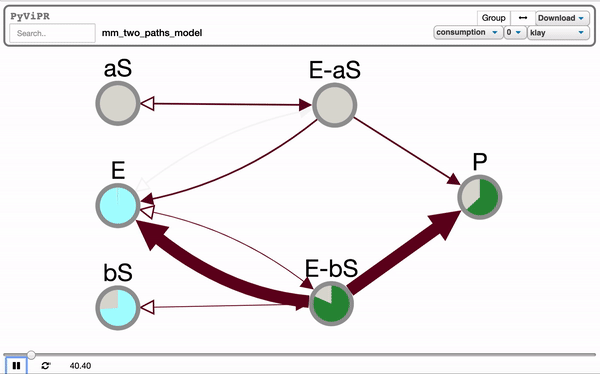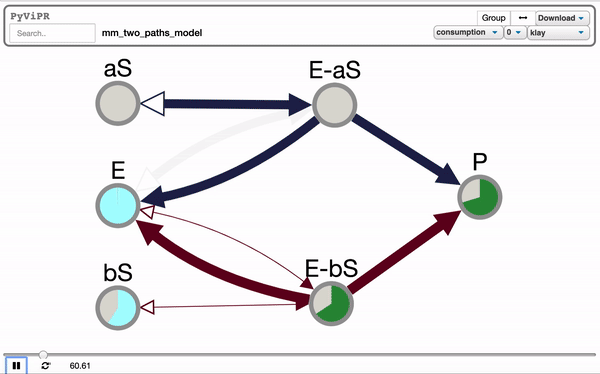
from pyvipr.examples_models.mm_two_paths_model import model
from pysb.simulator import ScipyOdeSimulator
import numpy as np
tspan = np.linspace(0, 1000, 100)
sim = ScipyOdeSimulator(model, tspan).run()
pviz.sp_dyn_view(sim)Tellurium is needed to visualize Tellurium models and it is needed if you want to use the pyvipr.tellurium_viz module:
Installing Tellurium from pip:
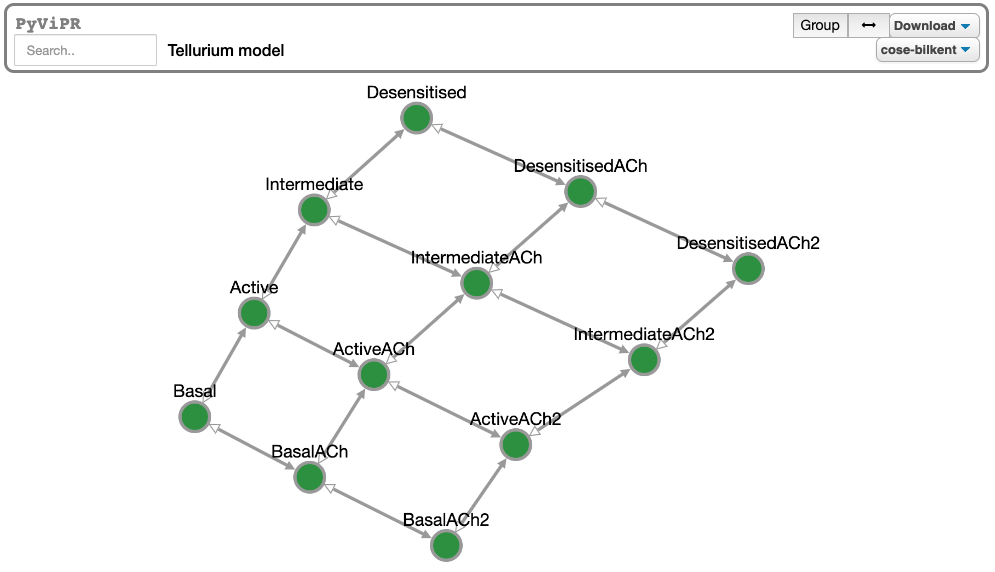
> pip install telluriumimport tellurium as te
import pyvipr.tellurium_viz as tviz
model = te.loadSBMLModel("https://www.ebi.ac.uk/biomodels-main/download?mid=BIOMD0000000001")
tviz.sp_view(model)To get started with using PyViPR, check out the full documentation
https://pyvipr.readthedocs.io/
To cite PyViPR, please cite the iScience paper:
Interactive Multiresolution Visualization of Cellular Network Processes
Ortega O and Lopez C